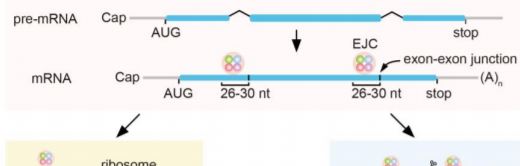
Telling Footprints: Exon Junction Complexes Mark Targets of Nonsense- and miRNA-Mediated mRNA Decay
RNA degradation mechanisms control the quality and quantity of mRNAs and are crucial for plant cell viability. The major pathways for RNA degradation involve 5’-3’ or 3’-5’ exonucleolytic cleavage by exoribonucleases or the exosome complex, respectively, whereas targets of miRNAs are cleaved by RNA endonucleases (Zhang and Guo, 2017). The exoribonuclease XRN4 degrades mRNA intermediates with free 5’ monophosphate (5’P) resulting from 5’ cap removal or endonucleolytic RNA cleavage. Proteins bound on mRNA can protect it from degradation, allowing the identification of their binding regions in mRNAs through footprinting approaches, such as ribosome profiling that is used to identify actively translated mRNAs. Similarly, exon junction complexes (EJCs) deposited upstream of the exon-exon junctions on mRNAs during splicing and removed upon the first round of translation, can protect mRNAs from degradation. In the current issue, Lee et al. (2020) show that the RNA degradome contains EJC footprints that mark mRNAs degraded during or before the pioneer round of translation. EJC footprints were prevalent in targets of nonsense-mediated decay (NMD) pathway, but were also present in some mRNAs degraded through miRNA-mediated cleavage.
Lee et al. (2020) analyzed RNA degradome data for 5’P end occurrence and found that it was relatively higher in the regions 25-30 nt upstream of the 3’ ends of exons, where EJCs are canonically positioned. This pattern was present in Arabidopsis, rice, human, and worm datasets, but not in budding yeast that lacks EJCs, suggesting that the 5’P peaks corresponded to 5’ termini of RNA degradation intermediates protected from degradation by EJCs. The xrn4 and fry1 mutants deficient in 5’-3’ RNA decay had less 5’P ends in the canonical EJC region, suggesting that EJCs hinder XRN-mediated 5’-3’ RNA degradation.
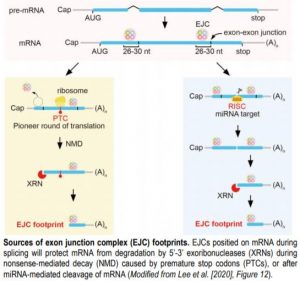 The authors also found a strong link between the NMD pathway and the EJC footprints, as a third of the genes regulated by the NMD pathway had enriched 5’P ends in the EJC region, whereas only ~15% of all intron-containing genes showed similar enrichment. During NMD, ribosomal arrest on a premature termination codon (PTC) caused by alternative splicing or mutation leads to decapping and 5’-3’ RNA decay, but EJCs on exons downstream of the PTC should protect the mRNA and lead to degradation intermediates with 5’P ends in the EJC region. In line with this, in several genes that produce mRNAs with PTCs due to alternative splicing, enrichment of 5’P peaks in the EJC regions of the exons 3’ to the PTC was found that was reduced in the xrn4 and fry1 mutants. Similarly, in plants with mutations leading to PTCs in mRNAs, most 5’ ends of the RNA degradation fragments mapped to the region corresponding to the canonical EJC binding region downstream of the PTCs, whereas the abundance of these footprints was lower, when the mutant plants were also deficient in XRN4 function.
The authors also found a strong link between the NMD pathway and the EJC footprints, as a third of the genes regulated by the NMD pathway had enriched 5’P ends in the EJC region, whereas only ~15% of all intron-containing genes showed similar enrichment. During NMD, ribosomal arrest on a premature termination codon (PTC) caused by alternative splicing or mutation leads to decapping and 5’-3’ RNA decay, but EJCs on exons downstream of the PTC should protect the mRNA and lead to degradation intermediates with 5’P ends in the EJC region. In line with this, in several genes that produce mRNAs with PTCs due to alternative splicing, enrichment of 5’P peaks in the EJC regions of the exons 3’ to the PTC was found that was reduced in the xrn4 and fry1 mutants. Similarly, in plants with mutations leading to PTCs in mRNAs, most 5’ ends of the RNA degradation fragments mapped to the region corresponding to the canonical EJC binding region downstream of the PTCs, whereas the abundance of these footprints was lower, when the mutant plants were also deficient in XRN4 function.
Although the EJC-footprints were mostly associated with NMD-mediated 5’-3’ RNA decay, they were found also downstream of some miRNA-guided RNA cleavage sites, whereas respective EJC-footprints were less abundant in the xrn4 and fry1 mutants, suggesting that XRN4 and other 5’-3’ exonucleases degrade the cleavage products of miRNA target mRNAs. The reduction of EJC footprints of some miRNA target transcripts in the ago1 mutant that is deficient in the key mediator of miRNA function, further supported the contribution of miRNA-mediated mRNA degradation to EJC footprint accumulation.
The study by Lee et al. (2020) indicates that EJC-footprints mark mRNAs that are degraded during or before the pioneer round of translation, many of them through the NMD pathway. Therefore, novel NMD targets could be identified through EJC footprinting. However, as EJC footprints were also associated with some miRNA targets, further research is required to clarify the sources of EJC footprints in the RNA degradome and to better understand the complex regulation of plant mRNA degradation.
Hanna Hõrak
Institute of Technology
University of Tartu, Estonia
ORCID: 0000-0002-6392-859X
REFERENCES
Lee, W.-C., Hou, B.-H., Hou, C.-Y., Tsao, S.-M., Kao, P. and Chen, H.-M. (2020). Widespread Exon Junction Complex Footprints in the RNA Degradome Mark mRNA Degradation Before Steady-state Translation. Published January 2020. DOI: https://doi.org/10.1105/tpc.19.00666.
Zhang, X. and Guo, H. (2017). mRNA decay in plants: both quantity and quality matter. Curr. Opin. Plant Biol. 35: 138–144.