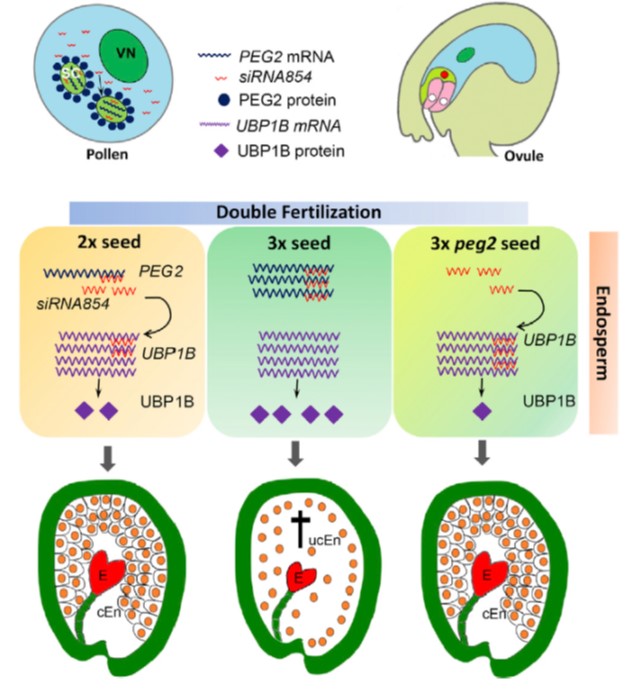
The imprinted gene PEG2 acts as a sponge for the transposon-derived siRNA854, inducing postzygotic reproductive isolation (Devel. Cell)
Plant Science Research WeeklyClosely related species that have different numbers of chromosomes (e.g., 2n versus 4n) are reproductively isolated, and this can arise as a consequence of an unbalancing in the expression levels of maternally- and paternally-imprinted genes. Wang et al. have identified a fascinating mechanism that explains…
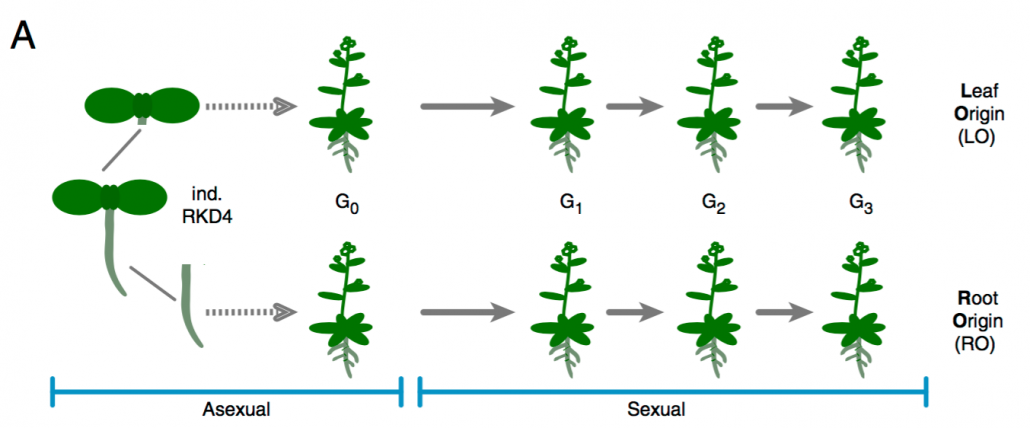
Heritable phenotypic variation due to partial maintenance of organ-specific epigenetic marks during asexual reproduction ($) (PNAS)
Plant Science Research WeeklyClonal propagation is widely used by humans to maintain and propagate desirable phenotypic traits in plants. Despite a restricted genetic variety, phenotypical difference, known a somaclonal variation, arises sometimes between parents and the clonally propagated progeny. Some of this variability is attributed…
Postzygotic reproductive isolation via sequestration of a transposon-derived siRNA ($) (Devel. Cell)
Plant Science Research WeeklyHybridization of plants with distinct chromosome number often results in seed development defects due to a phenomenon known as the triploid block. While this is triggered by the unbalanced expression of some paternally and maternally imprinted alleles, the molecular basis of the triploid block is not…
Stony hard phenotype in peach due to transposon insertion into YUCCA ($) (Plant J)
Plant Science Research WeeklyFruit softening in melting-flesh peaches is triggered by a major accumulation of ethylene at the late stage of ripening. Existence of stony hard peaches showing inhibition of fruit softening has been correlated with low levels of indole-3-acetic-acid inducing low levels of ethylene, but the underlying…
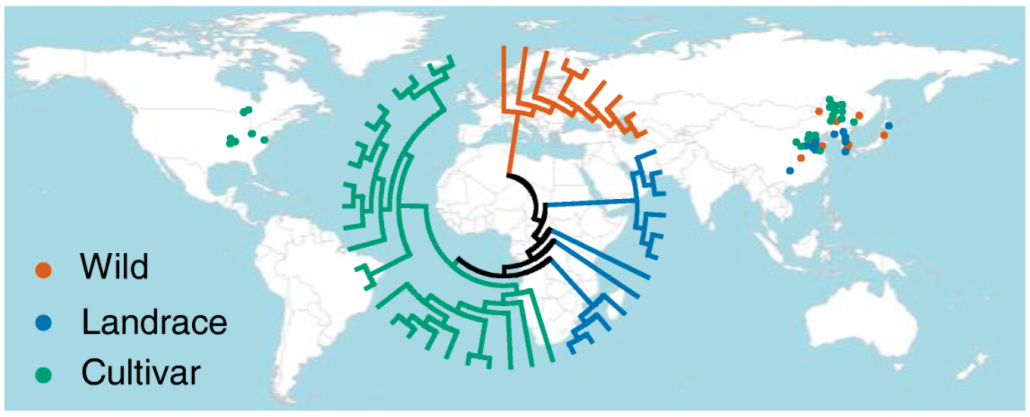
DNA methylation footprints during soybean domestication and improvement (Genome Biol)
Plant Science Research WeeklyCrop domestication relies on harnessing the natural genomic diversity present in the cultivated population. The contribution of genetic variation for selection and improvement of traits in plants has been extensively studied. Besides this, variation in epigenetic components such as DNA methylation offers…
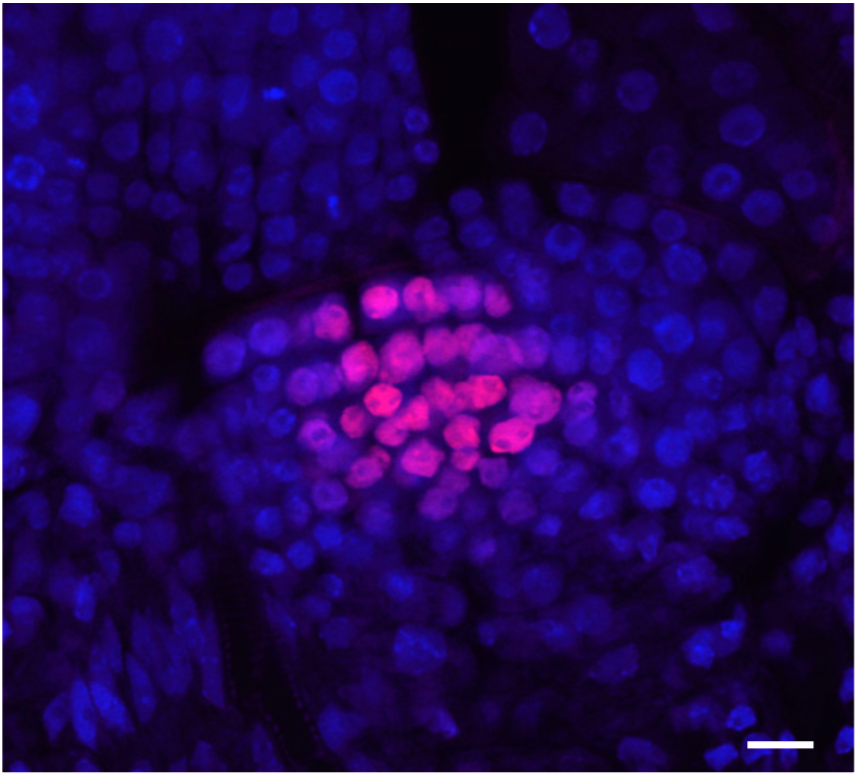
Early and transient loss of transposon control in Arabidopsis shoot stem cells (bioRxiv)
Plant Science Research WeeklyPostembryonic development in plants relies on stem cells located within the shoot apical meristem. Male and female gametes are descendants of those stem cells and maintenance of genome stability in this pool of cells is thus fundamental. In-depth molecular analysis has so far been hindered by the tedious…
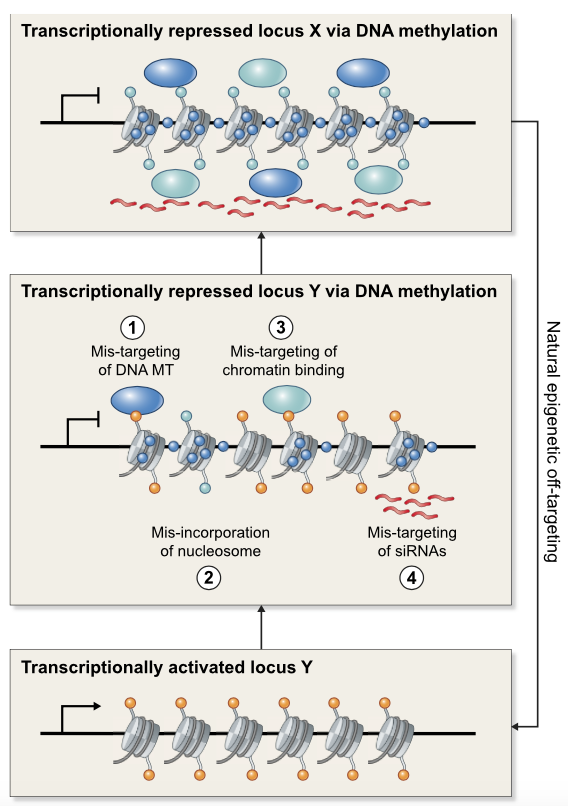
Review: Spontaneous epimutations in plants (New Phytol)
Plant Science Research WeeklySpontaneous epimutations are stochastic and heritable changes of DNA methylation in genomes. In this review, Johannes and Schmitz discuss the molecular origins of spontaneous epimutations in plants as well as their functional and phenotypical consequences. The authors highlight that spontaneous epimutations…
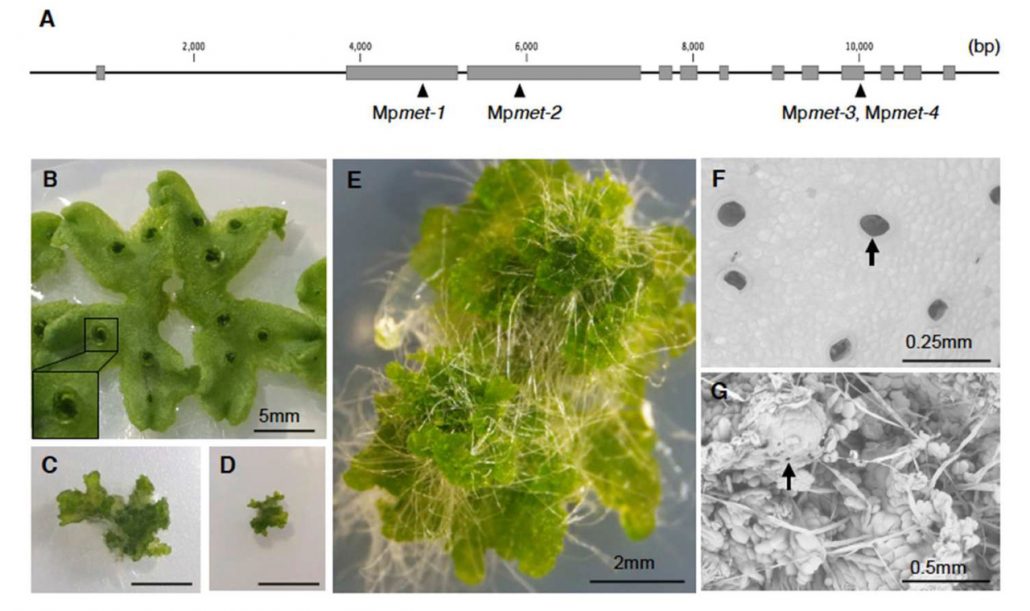
Non-CG methylation mechanism in Marchantia polymorpha ($) (Plant Cell Physiol.)
Plant Science Research WeeklyPrevious reports have shown that M. polymorpha DNA methylation status varies during its life cycle and it contains only one copy of the METHYLTRANSFERASE1 (MET1) orthologous gene. MET1 is responsible of CG methylation in angiosperms. Ikeda and collaborators generated MpMET mutants using the CRISPR/CAS9…
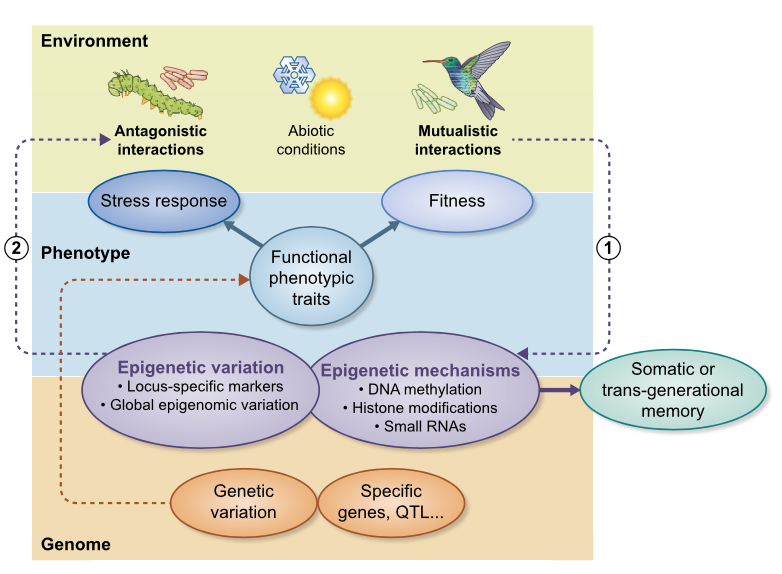
Review: The role of plant epigenetics in biotic interactions (New Phytol.)
Plant Science Research WeeklyPlant phenotypes are influenced by the nature and intensity of biotic interactions. While the role of genetic diversity has been extensively studied, contribution of epigenetics to plant fitness and response to biotic stresses remains elusive. The authors review here the most recent literature about…

