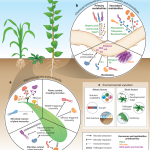
Sulfotransferase 1 is the enzymatic hub of sulfated salicinoids in poplar.
Plants produce a plethora of specialized metabolites serving as a chemical arsenal against abiotic and biotic challenges during their lifetime. Core metabolites serve as building blocks for diverse decorating enzymes like oxidases, reductases or transferases that introduce new functionalities expanding the structural diversity and resulting in an astonishing metabolic palette dispersed in the plant kingdom. Some enzymatic hubs are not broadly distributed but are limited in specific families, genera or even species, resulting in taxon-restricted compounds. One example of decoration is the sulfation of secondary compounds, an enzymatic reaction performed by members of the sulfotransferase family (SOTs, EC 2.8.2.-) (Günal et al., 2019). Known types of sulfated metabolites are – among others – flavonoids, glucosinolates and terpenoids but only in few cases the corresponding SOT enzyme has been identified (Hirschmann et al., 2014).
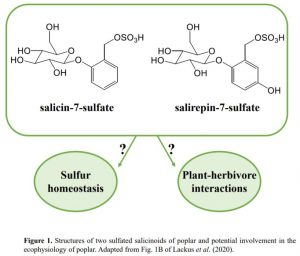 In this issue of Plant Physiology, Lackus et al. (2020) expand the list of known sulfated compounds by identifying two sulfated salicinoids in poplar. Salicinoids are a sub-class of phenolic glycosides characteristic of the Salicaceae family and serve as defense compounds in planta against herbivores (Boeckler et al., 2011). The authors used LC-MS to compare the spectra of methanolic extracts from leaves of two salicinoid-abundant species (Populus trichocarpa and Populus x canescens) with a low-salicinoid-accumulating plant (Salix viminalis). NMR analysis confirmed the structure of salicin-7-sulfate, a compound already described in Salix sp. (Noleto-Dias et al., 2018), and salirepin-7-sulfate, which is a previously undescribed compound (Fig. 1). Both of these compounds were present in a broad spectrum of Salicaceae species, and mostly accumulated in the aerial organs. Among the screened species in Salicaceae, the authors identified two exceptions that accumulated either only salicin-7-sulfate (S. caprea) or none of the sulfated salicinoids (P. nigra and S. viminalis).
In this issue of Plant Physiology, Lackus et al. (2020) expand the list of known sulfated compounds by identifying two sulfated salicinoids in poplar. Salicinoids are a sub-class of phenolic glycosides characteristic of the Salicaceae family and serve as defense compounds in planta against herbivores (Boeckler et al., 2011). The authors used LC-MS to compare the spectra of methanolic extracts from leaves of two salicinoid-abundant species (Populus trichocarpa and Populus x canescens) with a low-salicinoid-accumulating plant (Salix viminalis). NMR analysis confirmed the structure of salicin-7-sulfate, a compound already described in Salix sp. (Noleto-Dias et al., 2018), and salirepin-7-sulfate, which is a previously undescribed compound (Fig. 1). Both of these compounds were present in a broad spectrum of Salicaceae species, and mostly accumulated in the aerial organs. Among the screened species in Salicaceae, the authors identified two exceptions that accumulated either only salicin-7-sulfate (S. caprea) or none of the sulfated salicinoids (P. nigra and S. viminalis).
BLAST searching the previously characterized salicylic acid sulfotransferase (AtSOT12) from Arabidopsis against the genome of P. trichocarpa resulted in the identification of 28 candidates that potentially encode SOTs. Phylogenetic and transcriptomic analysis of the candidates indicated that only two were highly expressed in the leaves of poplar, where sulfated salicinoids mostly accumulate. Heterologous expression followed by in vitro assays with the purified enzymes revealed that one of the two candidates (PtSOT1) could convert salicin and salirepin into the respective sulfated forms. RT-qPCR analysis of PtSOT1 further confirmed that the abundance of transcripts was in agreement with the spatial accumulation of sulfated salicinoids in various organs.
As a demonstration of the function of SOT1, the authors established that the absence of sulfated salicinoids in P. nigra is caused by a frame-shift mutation in the genomic region of PnSAT1 that results in a non-functional enzyme. In agreement with this natural mutation, RNAi-mediated silencing of the SOT1 transcripts in P. x canescens resulted in significantly lower levels of sulfated salicinoids compared to the controls, indicating that SOT1 is necessary and sufficient for the biosynthesis of salicin-7-sulfate and salirepin-7-sulfate in planta.
To tease out a possible role of sulfated salicinoids against herbivores, the authors conducted feeding experiments with caterpillars of the generalist feeder Lymantria dispar. No difference in the levels of salicin-7-sulfate and salirepin-7-sulfate could be observed in response to herbivory. Furthermore, sulfated salicinoids most likely do not serve as feeding deterrents, since no feeding preference could be determined for L. dispar among leaves of either wild-type plants or the SOT1 RNAi plants. Further research with specialized feeding insects like the red poplar leaf beetle (Chrysomela populi) that have evolved mechanisms to cope with salicinoids (Michalski et al., 2008) could possibly reveal a role for sulfated salicinoids in plant–herbivore interactions.
Elemental analysis in poplar leaves showed that almost 5.5% of the total sulfur and 7.5% of free and hydrolysable sulfate are represented as sulfated salicinoids, suggesting that salicin-7-sulfate and salirepin-7-sulfate represent a strategy to store sulfate and thus contribute to sulfur homeostasis. This result is in close agreement with the amount of sulfur in Brassicaceae species tied up in glucosinolates, which have been suggested to serve as a sulfur-storage pool (Falk et al., 2007). Since the habitats of poplar and willow trees are wetland or riparian soils, sulfation of salicinoids might have been recruited to cope with low sulfate availability due to flooding. Some genotypes of P. nigra are drought tolerant (Viger et al., 2016); therefore it would be interesting to address whether the lack of sulfated salicinoids in P. nigra could potentially point to an evolutionary trade-off between biosynthesis of sulfated salicinoids and drought tolerance.
Although a wide range of functions has been assigned to sulfated metabolites, few SOTs have been characterized in detail. The work of Lackus et al. (2020) broadens the list of sulfated compounds and most importantly couples it with the functional characterization of the corresponding sulfotransferase.
Konstantinos Koudounas, Assistant Features Editor
ORCID:0000-0002-6000-6565
EA2106 – Biomolecules and Plant Biotechnology
University of Tours
Tours, 37200 France
References
Boeckler GA, Gershenzon J, Unsicker SB (2011) Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 72: 1497-1509
Falk KL, Tokuhisa JG, Gershenzon J (2007) The Effect of Sulfur Nutrition on Plant Glucosinolate Content: Physiology and Molecular Mechanisms. Plant biol (Stuttg) 9: 573-581
Günal S, Hardman R, Kopriva S, Mueller JW (2019) Sulfation pathways from red to green. Journal of Biological Chemistry 294: 12293-12312
Hirschmann F, Krause F, Papenbrock J (2014) The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Frontiers in Plant Science 5: 556
Lackus ND, Müller A, Kröber TDU, Reichelt M, Schmidt A, Nakamura Y, Paetz C, Luck K, Lindroth RL, Constabel CP, Unsicker SB, Gershenzon J, Köllner TG (2020) The occurrence of sulfated salicinoids in poplar and their formation by sulfotransferase 1. Plant Physiology: pp.01447.02019 https://doi.org/10.1104/pp.19.01447
Michalski C, Mohagheghi H, Nimtz M, Pasteels J, Ober D (2008) Salicyl Alcohol Oxidase of the Chemical Defense Secretion of Two Chrysomelid Leaf Beetles: Molecular and functionalcharacterization of two new members of the glucose-methanol-choline oxidoreductase gene family. Journal of Biological Chemistry 283: 19219-19228
Noleto-Dias C, Ward JL, Bellisai A, Lomax C, Beale MH (2018) Salicin-7-sulfate: A new salicinoid from willow and implications for herbal medicine. Fitoterapia 127: 166-172
Viger M, Smith HK, Cohen D, Dewoody J, Trewin H, Steenackers M, Bastien C, Taylor G (2016) Adaptive mechanisms and genomic plasticity for drought tolerance identified in European black poplar (Populus nigra L.). Tree Physiology 36: 909-928