Stress Management: OsIDS1 Modulates Histone Deacetylation to Repress Salt Tolerance Genes
Plants evolved to react to the changing environment by developing sensing and signal transduction mechanisms, which ultimately lead to changes in gene expression and altered plant performance. Transcription factors (TFs) are key to regulating the expression of individual genes. TFs can confer sequence specificity to chromatin remodelers, further shaping the transcriptional landscape in response to a changing environment. Although Barbara McClintock herself recognized the importance of epigenetic regulation in stress response (McClintock, 1984), how epigenetic changes arise in response to stress still is not fully understood.
Histone acetylation is one of the best-studied histone modifications (Lawrence et al., 2016). Positively charged Lys residues of the histone tails bind to negatively charged DNA, resulting in a tight chromatin structure (Verdone et al., 2006). Those Lys residues often are targets of histone acetylases. Histone acetylation neutralizes the positive charge of Lys residues and reduces nucleosome binding, resulting in a more accessible chromatin structure. Acetyl groups are removed by histone deacetylates (HDAs) that, like histone acetylases, lack intrinsic DNA-binding activity. HDAs are recruited to their target sites via association with sequence-specific transcriptional repressors, which possess motifs contributing to transcriptional repression (Kagale and Rozwadowski, 2011). One of the first repression-associated motifs identified in plants is the EAR motif (for ethylene response factor-associated amphiphilic repression; Ohta et al., 2001). The EAR motif is responsible for interactions with other repressor proteins, including TOPLESS/Topless-like (TPL/TPR; Ke et al., 2015), for attracting HDAs and resulting in inaccessible chromatin structure. The EAR motif is so effective in transcriptional repression that when it is fused to a transcriptional activator, the hybrid TF turns into a dominant repressor (Hiratsu et al., 2003).
In this issue of Plant Physiology, Cheng et al. (2018) screened the rice (Oryza sativa) genome for genes encoding TFs containing EAR motifs. Out of the total of 374 genes encoding EAR-TFs, the group further identified 57 genes that showed a change in expression in response to salt stress. These 57 TF genes were individually targeted by artificial microRNA interference. Rice seedlings stably transformed with the artificial microRNA constructs were screened for changes in yield and survival under salt stress. Among the 20 TFs with enhanced or decreased salt stress tolerance, the authors identified indeterminate spikelet 1 (OsIDS1), previously reported to be involved in regulation of floral meristem architecture (Lee and An, 2012). Reduced expression of OsIDS1 resulted in increased salinity tolerance, while the opposite was observed in OsIDS1 overexpression lines, suggesting that IDS1 is a negative regulator of salt tolerance. ChIP-Seq studies indicated that IDS1 predominantly targets promoter regions of genes involved in plant development and responses to abiotic stress, for example, late embryogenesis abundant 1 (OsLEA1) and salt overly sensitive 1 (OsSOS1). Yeast two-hybrid studies identified TPR1 and HDA1 as proteins that interact with IDS1. Based on their results, the authors propose that TPR1/HDA1/IDS1 form a transcriptional repression protein complex in vivo that antagonizes access of RNA Polymerase II to IDS1-targeted genes, through histone deacetylation.
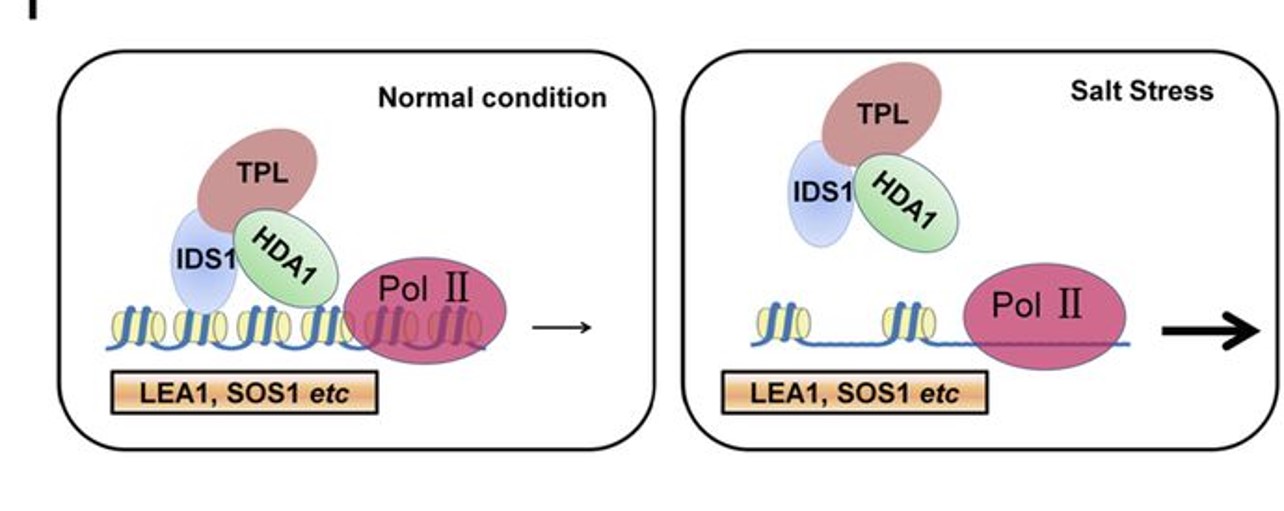 The work by Cheng et al. (2018) provides a detailed insight into gene regulation through epigenetic modifications. Understanding the upstream mechanisms controlling OsIDS1 expression might shed light on how chromatin structure is altered under salt stress. Reducing the expression of stress-related genes seems compelling under optimal growth conditions as it could prevent the plant from using its resources for unnecessary stress response. The finding that IDS1 expression is up-regulated by salt treatment yet IDS1 serves to repress salt-responsive genes is puzzling. In floral development, IDS1 transcript levels are regulated by miRNA172 (Lee and An, 2012), while miRNA172 itself is regulated by light through the phytochromes (Lee et al., 2014). Whether this control unit also is involved in modulating stress responses remains to be established. Identification of molecular components acting upstream of OsIDS1 under salt stress will reveal how the TPR1/HDA1/IDS1 complex is affected during salt stress exposure, potentially revealing a new signal-relay mechanism.
The work by Cheng et al. (2018) provides a detailed insight into gene regulation through epigenetic modifications. Understanding the upstream mechanisms controlling OsIDS1 expression might shed light on how chromatin structure is altered under salt stress. Reducing the expression of stress-related genes seems compelling under optimal growth conditions as it could prevent the plant from using its resources for unnecessary stress response. The finding that IDS1 expression is up-regulated by salt treatment yet IDS1 serves to repress salt-responsive genes is puzzling. In floral development, IDS1 transcript levels are regulated by miRNA172 (Lee and An, 2012), while miRNA172 itself is regulated by light through the phytochromes (Lee et al., 2014). Whether this control unit also is involved in modulating stress responses remains to be established. Identification of molecular components acting upstream of OsIDS1 under salt stress will reveal how the TPR1/HDA1/IDS1 complex is affected during salt stress exposure, potentially revealing a new signal-relay mechanism.
Although IDS1 is an appealing target for stress tolerance breeding, this road to enhanced stress tolerance might be a bumpy one. For example, releasing the transcription of stress-related genes might lead to an undesired restriction of growth, limiting yield. Moreover, previous studies show that epigenetic regulators have different effects depending on the genetic context. For example, mutations affecting AtHDA19 expression increased salt stress tolerance in the Arabidopsis Ws accession but reduced salt tolerance in the Col-0 accession (Chen and Wu, 2010; Mehdi et al., 2015). Although Cheng et al. (2018) showed that IDS1 acts as a negative regulator of salt tolerance in the O. sativa japonica cv Nipponbare, its effect remains to be established in other genetic backgrounds, such as indica varieties, which commonly are grown in salt-affected regions of South East Asia.
REFERENCES
Chen L-T, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5: 1318–1320
Cheng X, Zhang S, Tao W, Zhang X, Liu J, Sun J, Zhang H, Pu L, Huang R, Chen T (2018) INDETERMINATE SPIKELET1 recruits histone deacetylase and a transcriptional repression complex to regulate rice salt tolerance. Plant Physiol 178:
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739
Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146
Ke J, Ma H, Gu X, Thelen A, Brunzelle JS, Li J, Xu HE, Melcher K (2015) Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci Adv 1: e1500107
Lawrence M, Daujat S, Schneider R (2016) Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32: 42–56
Lee D-Y, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461
Lee Y-S, Lee D-Y, Cho L-H, An G (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice (N Y) 7: 31
McClintock B (1984) The significance of responses of the genome to challenge. Science 226: 792–801
Mehdi S, Derkacheva M, Ramström M, Kralemann L, Bergquist J, Hennig L (2015) The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28: 42–54
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968
Verdone L, Agricola E, Caserta M, Di Mauro E (2006) Histone acetylation in gene regulation. Brief Funct Genomics Proteomics 5: 209–221