Polycomb Group Transcriptional Repressor: Suppress to Sustain
Evidence of epigenetic gene regulation in plant immunity was first reported in 1975 (Guseinov et al., 1975) with the demonstration that cytosine methylation is altered in response to pathogen infection. Since then, it has been established that pathogen infection influences DNA methylation and histone modification patterns. In addition, histone methyl transferase and demethylases have been described as immunity regulators, with their mutants displaying compromised resistance (Ramirez-Prado et al., 2018). Previously, it was shown that Arabidopsis (Arabidopsis thaliana) plants exposed to the bacterial pathogen Pseudomonas syringae (Pst) rapidly altered its methylated cytosine pattern, which in turn contributed to the modulation of their defense response (Dowen et al., 2012).
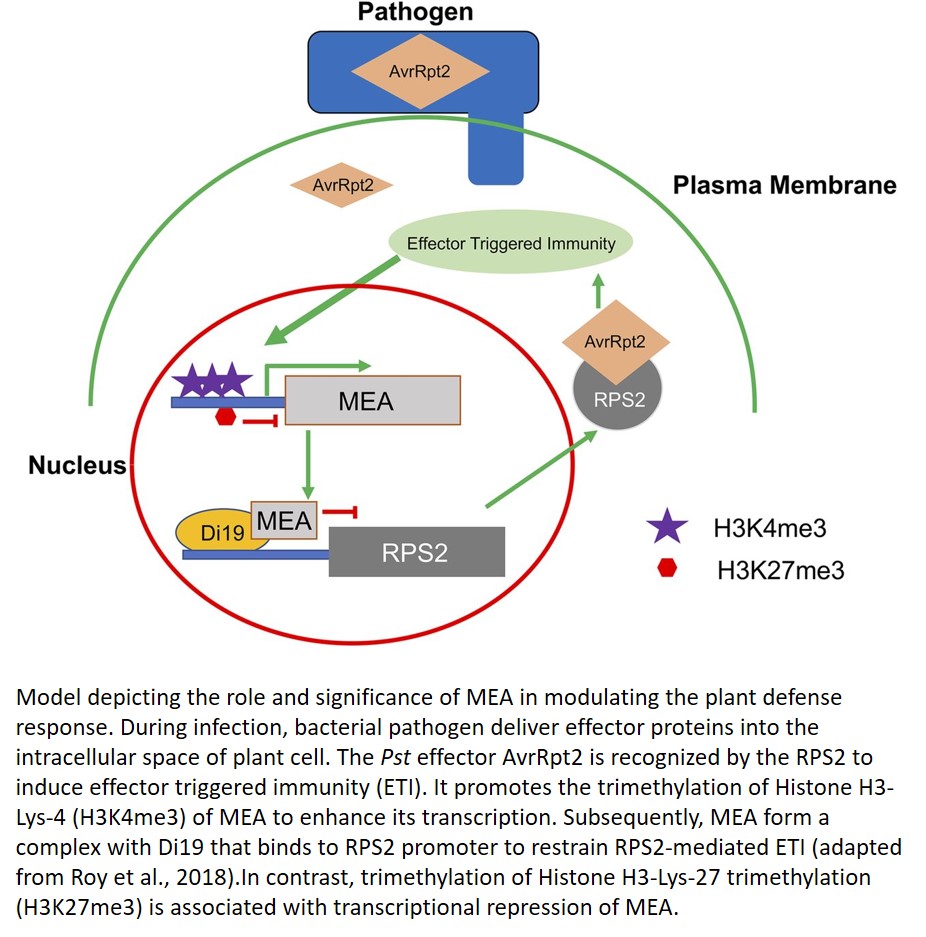
Roy et al. (2018) now find important overlaps between a Polycomb Group (PcG) transcriptional repressor and immune responses during pathogen infection. They identify a previously unknown PcG-mediated differential immunity response of Arabidopsis to virulent (Pst DC3000) and avirulent (Pst-AvrRpt2) pathogens. The authors observed delayed expression of MEDEA (MEA) after infection by the virulent pathogen, in contrast to a high level of MEA transcript accumulation within 6 h of avirulent pathogen infection, using MEA promoter-driven GUS (MEA:uidA) as a reporter in transgenic plants. MEA expression also was enhanced after salicylic acid and jasmonic acid treatments. These data indicate that the PcG core complex component MEA is induced during defense activation and contributes to modulation of effector-triggered immunity (ETI).
Although various candidate genes are known to be involved in the regulation of growth and immunity in plants independently, the role of these genes in the growth-immunity trade-off during pathogen infection is yet to be understood. PcG protein complexes are conserved in plants and animals and function as transcriptional repressors by limiting transcription factor access to chromatin through modification of histones. In plants, PcG proteins are known to regulate female gametophyte and seed development. The highly conserved Polycomb Repressive Complex2 (PRC2) represses thousands of target genes by mono-, di-, and trimethylation of Histone H3 on Lys-27 (Pengelly et al., 2013) as a key epigenetic regulator of chromatin modification. The maternally expressed MEA belongs to the PRC2 group and confers maternal control over seed development as well as represses expression of downstream target genes (Schubert et al., 2005). This study establishes a role for MEA, a tightly controlled developmental gene, in attenuating defense responses in plants, thereby offering a balance between growth and defense in plants.
Since the homozygous mea-6 mutation is embryo lethal, Roy et al. used the elegant centromere-mediated chromatin elimination method (Ravi and Chan, 2010) to generate a mea-6 haploid that was converted to a homozygous mea-6 diploid for further analysis. Although a majority of seeds from the double haploid mea-6 plants were inviable, of those that were viable only 30% displayed delayed germination relative to the wild type. These mea-6 mutants were resistant to both virulent and avirulent strains of Pst. By contrast, pro35S:MEA transgenic overexpression lines showed reduced immunity to Pst infection and a reduced hypersensitive response as measured by ion leakage. Surprisingly, MEA overexpression plants had elevated disease symptoms and lower levels of PLANT DEFENSIN1.2 transcripts than wild-type plants when challenged with the necrotrophic pathogen Botrytis cinerea. These data indicate a potential role of MEA as a negative feedback regulator of defense as well as its role in maternal gametophyte development.
Roy et al. (2018) also explored the mechanism behind the expression of MEA during Pst-AvrRpt2 infection. They report that the Leu-rich repeat class receptor RPS2 is required for Pst-AvrRpt2-mediated MEA expression. Furthermore, chromatin immunopurification-PCR analyses implicated MEA in RPS2 transcriptional regulation. In addition, authors performed an Arabidopsis cDNA library screen using MEA as a bait and identified a nucleus-localized zinc finger transcriptional activator, DROUGHT INDUCED19 (Di19/AtDi19-1), as an interacting partner (Fig. 1). Di19 is well known to bind to the promoters and regulate the expression of the PATHOGENESIS-RELATED1 (PR1), PR2, and PR5 genes (Liu et al., 2013). Interestingly, the authors have identified a Di19 binding site in the promoter region of RPS2. They were further able to show that MEA and Di19 form a necessary complex to regulate RPS2 expression.
In short, Roy et al. (2018) highlight the significance of the PcG protein MEA and bridge the gap between regulatory processes associated with development and gametogenesis and those associated with pathogen defense and plant immunity. This knowledge may offer new strategies for manipulation of defense traits without compromising yield.
REFERENCES
Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109: E2183–E2191
Guseinov VA, Kiryanov GI, Vanyushin BF (1975) Intragenome distribution of 5-methylcytosine in DNA of healthy and wilt-infected cotton plants (Gossypium hirsutum L.). Mol Biol Rep 2: 59–63
Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH, Chen YF (2013) Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant 6: 1487–1502
Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J (2013) A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699
Ramirez-Prado JS, Piquerez SJM, Bendahmane A, Hirt H, Raynaud C, Benhamed M (2018) Modify the histone to win the battle: chromatin dynamics in plant-pathogen interactions. Front Plant Sci 9: 355
Ravi M, Chan SW (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618
Roy S, Gupta P, Rajabhoj MP, Maruthachalam R, Nandi AK (2018) The polycomb-group repressor MEDEA attenuates pathogen defense. Plant Physiol 177: 1728–1742
Schubert D, Clarenz O, Goodrich J (2005) Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 8: 553–561