Stop the FUSS: BPCs restrict FUSCA3 transcription to promote ovule and seed development
Tightly controlled genetic programs regulate developmental phase transitions within distinct tissues and cells. This is especially true for the vegetative-to-reproductive and reproductive-to-seed developmental phase changes that ensure the production of gametes capable of generating viable seed upon fertilization. The LAFL transcription factors LEC2 (LEAFY COTYLEDON2), ABI3 (ABA INSENSITIVE3), FUS3 (FUSCA3), and LEC1 (LEAFY COTYLEDON1) have emerged as master regulators of seed development in angiosperms. These genes are sequentially expressed and act in concert to promote embryo development and seed maturation while also preventing early germination and vegetative growth (Lepiniec et al., 2018).
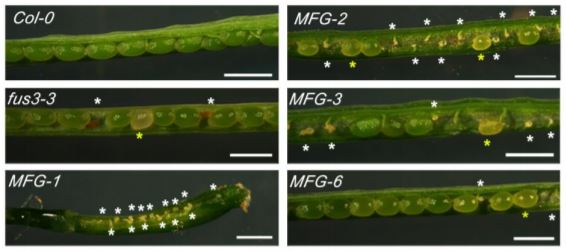
 The LAFL transcription factor FUS3 plays an important role in reproductive development, as null fus3-3 mutants exhibit enhanced seed abortion phenotypes under elevated temperatures and FUS3 mis-expression lines display aborted siliques (Gazzarrini et al., 2004; Chan et al., 2017). To further explore this paradigm, Wu et al. (2020) examined the spatiotemporal expression and function of FUS3 during reproductive stages in Arabidopsis thaliana. Using a stable, non-functional FUS3pro:FUS3ΔC-GFP reporter construct, the authors observed FUS3ΔC-GFP expression within the pistil and ovules of Arabidopsis flowers. During ovule development GFP accumulation was observed in the nucellus epidermis, chalaza, and funiculus, whereas mature ovules only exhibited GFP within the chalaza and funiculus. The importance of FUS3 expression within these domains was determined by monitoring ovule development in fus3-3 mutant and ML1 (MERISTEM LAYER1) promoter driven FUS3-GFP mis-expression lines. Compared to wildtype plants, both fus3-3 and ML1:FUS3-GFP lines exhibited embryo sacs that were either delayed, arrested, or malformed (not wrapped by integuments), ultimately resulting in ovule abortion (Figure).
The LAFL transcription factor FUS3 plays an important role in reproductive development, as null fus3-3 mutants exhibit enhanced seed abortion phenotypes under elevated temperatures and FUS3 mis-expression lines display aborted siliques (Gazzarrini et al., 2004; Chan et al., 2017). To further explore this paradigm, Wu et al. (2020) examined the spatiotemporal expression and function of FUS3 during reproductive stages in Arabidopsis thaliana. Using a stable, non-functional FUS3pro:FUS3ΔC-GFP reporter construct, the authors observed FUS3ΔC-GFP expression within the pistil and ovules of Arabidopsis flowers. During ovule development GFP accumulation was observed in the nucellus epidermis, chalaza, and funiculus, whereas mature ovules only exhibited GFP within the chalaza and funiculus. The importance of FUS3 expression within these domains was determined by monitoring ovule development in fus3-3 mutant and ML1 (MERISTEM LAYER1) promoter driven FUS3-GFP mis-expression lines. Compared to wildtype plants, both fus3-3 and ML1:FUS3-GFP lines exhibited embryo sacs that were either delayed, arrested, or malformed (not wrapped by integuments), ultimately resulting in ovule abortion (Figure).
Since the spatiotemporal restriction of FUS3 is critical for ovule and early seed development, the authors performed yeast one-hybrid screening to identify transcriptional regulators of FUS3. By screening a library of transcription factors against a 615 base-pair fragment of the FUS3 promoter, Wu and colleagues identified class I BPC (BASIC PENTACYSTEINE) repressor proteins (BPC1-3) as candidate FUS3 regulators. Targeted promoter binding assays further demonstrated that BPCs bind to (GA/CT)n cis-elements present in the 5’ untranslated and the first intron/exon regions of FUS3 during reproductive development.
Previous research implicated BPCs in a wide range of developmental processes, as higher order bpc mutants exhibit ovule and seed abortion phenotypes (Monfared et al., 2011). Moreover, BPCs interact with PRC2 (POLYCOMB REPRESSOR COMPLEX2) subunits that were shown act at the FUS3 regions bound by class I BPCs (Xiao et al., 2017; Wu et al., 2020). To explore this further, the authors first confirmed that BPC1-3 interact with the PRC2 subunits FIS2 (FERTILIZATION INDEPENDENT SEED2) and MEDEA through yeast 2-hybrid and bimolecular fluorescence complementation assays. Expression analysis of BPC and FIS-PRC2 subunits throughout ovule and early seed development further supported the idea that these proteins mediate FUS3 repression during key phase transitions.
Since the morphology of bpc mutants largely resembles that of ML1pro-driven FUS3-GFP mis-expression lines, the authors hypothesized that FUS3 de-repression may contribute to these phenotypes. In support of this idea, expression analyses revealed ectopic and enhanced levels of FUS3 in bpc mutants relative to wildtype plants. Moreover, the fus3-3 mutation partially rescued the stunted growth, aborted embryo, and arrested seed phenotypes of bpc1/2 double mutants. Collectively, Wu et al. (2020) demonstrate that BPC proteins coordinate ovule and early seed development by recruiting PRC2 complexes that restrict FUS3 expression in key tissues and cell-types.
Philip Carella
Sainsbury Laboratory
University of Cambridge, Cambridge
ORCID: 0000-0002-5467-7290
REFERENCES
Chan A, Carianopol C, Tsai AY-L, Varatharajah K, Chiu RS and Gazzarrini, S (2017). SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. . Exp. Bot. 68: 4219–4231.
Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M and McCourt P (2004). The Transcription Factor FUSCA3 Controls Developmental Timing in Arabidopsis through the Hormones Gibberellin and Abscisic Acid. Developmental Cell 7: 373–385.
Lepiniec L, Devic M, Roscoe TJ, Bouyer D, Zhou DX, Boulard C, Baud S and Dubreucq B (2018). Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 31: 291–307.
Monfared MM, Simon MK, Meister RJ, Roig-Villanova I, Kooiker M, Colombo L, Fletcher JC and Gasser CS (2011). Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis: Roles of BASIC PENTACYSTEINE genes. Plant J. 66: 1020–1031.
Xiao J et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49: 1546–1552.