Spatially resolved, single-cell multi-omics atlas of soybean development
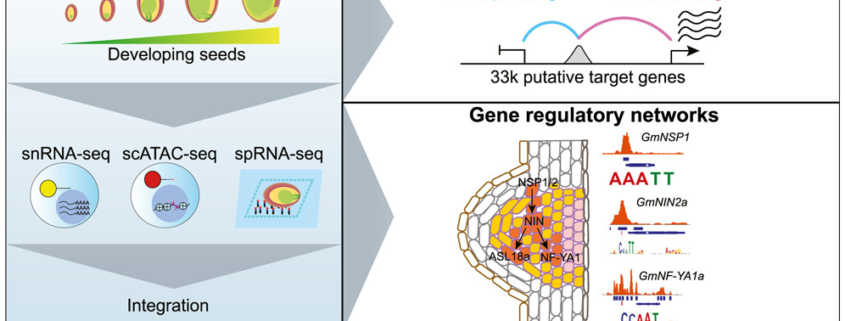
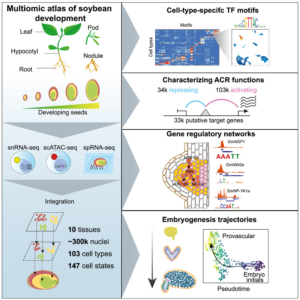 In this exciting paper, Zhang, Luo, Marand et al. combined several powerful techniques to investigate the program underlying soybean seed development. They used RNA sequencing to profile gene expression from single nuclei (snRNA-seq). They also carried out single-cell sequencing of assays for transposase-accessible chromatin (scATAC-seq) to identify accessible chromatin regions (ACR), which are potentially regions where transcription factors can bind. Combining these datasets with laser-capture microdissection RNA-seq datasets allowed the authors to pinpoint atlas-scale cell-type regulatory regions and gene expression profiles across 10 developmental stages and tissues. In addition to sharing these datasets in the Soybean Multi-Omics Atlas (https://soybean-atlas.com/), the authors share the first of surely many new insights. For example, they identified subtypes of endosperm cells with distinct patterns of expression of nutrient transporters and cell death, indicating a temporal as well as spatial pattern of nutrient support of the developing embryo. The analysis of the accessible chromatin regions (ACRs) is particularly interesting, as the authors found a high cell-type specific correlation between gene accessiblity and gene expression, and identified key transcription factors that define cell types; perhaps not surprising, but nevertheless intriguing. As the authors state, “We anticipate that the real potential of single-cell methods will extend beyond aiding gene function studies and uncovering regulatory networks.” Clearly these data provide a wealth of information to explore and analyze. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Cell 10.1016/j.cell.2024.10.050
In this exciting paper, Zhang, Luo, Marand et al. combined several powerful techniques to investigate the program underlying soybean seed development. They used RNA sequencing to profile gene expression from single nuclei (snRNA-seq). They also carried out single-cell sequencing of assays for transposase-accessible chromatin (scATAC-seq) to identify accessible chromatin regions (ACR), which are potentially regions where transcription factors can bind. Combining these datasets with laser-capture microdissection RNA-seq datasets allowed the authors to pinpoint atlas-scale cell-type regulatory regions and gene expression profiles across 10 developmental stages and tissues. In addition to sharing these datasets in the Soybean Multi-Omics Atlas (https://soybean-atlas.com/), the authors share the first of surely many new insights. For example, they identified subtypes of endosperm cells with distinct patterns of expression of nutrient transporters and cell death, indicating a temporal as well as spatial pattern of nutrient support of the developing embryo. The analysis of the accessible chromatin regions (ACRs) is particularly interesting, as the authors found a high cell-type specific correlation between gene accessiblity and gene expression, and identified key transcription factors that define cell types; perhaps not surprising, but nevertheless intriguing. As the authors state, “We anticipate that the real potential of single-cell methods will extend beyond aiding gene function studies and uncovering regulatory networks.” Clearly these data provide a wealth of information to explore and analyze. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Cell 10.1016/j.cell.2024.10.050