Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems (Nature Comms)
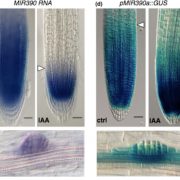
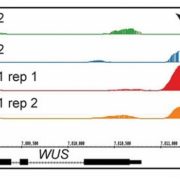
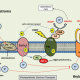
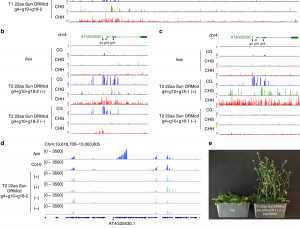 Steve Jacobsen’s group has been developing a broad range of synthetic biology tools to interrogate DNA-methylation in plants (see Gallego-Bartolomé et al., 2018, and Gallego-Bartolomé et al., 2019). Among them, they use the CRISPR-Cas9 SunTag system, in which the dCas9 protein is fused to SunTag, a repetitive peptide epitope. In combination with a guide RNA, this system allows for antibody-based recruitment of multiple copies of effectors. In this work by Papikian et al., they use it to recruit the VP64 transcriptional activator for locus-specific activation, or the DRM methyltransferase for locus-specific DNA-methylation. Although this method can increase the target gene’s transcript levels, it is not always enough to trigger a significant phenotype. In the same way, targeted DNA-methylation also initially presented some limitations of specificity and inheritability. However, the authors provide ways to overcome these problems and achieve desirable outputs (e.g., more gRNA, or by altering SunTag linker length, or effector nuclear localization signals). This paper is a good example of how a thorough analysis within a design-build-test-learn cyclic framework helps to the understanding of a technology and the observation of interesting phenomena. CRISPR-Cas9-SunTag system is a versatile expansion to the plant CRISPR toolbox. (Summary by Facundo Romani) Nature Comms. 10.1038/s41467-019-08736-7
Steve Jacobsen’s group has been developing a broad range of synthetic biology tools to interrogate DNA-methylation in plants (see Gallego-Bartolomé et al., 2018, and Gallego-Bartolomé et al., 2019). Among them, they use the CRISPR-Cas9 SunTag system, in which the dCas9 protein is fused to SunTag, a repetitive peptide epitope. In combination with a guide RNA, this system allows for antibody-based recruitment of multiple copies of effectors. In this work by Papikian et al., they use it to recruit the VP64 transcriptional activator for locus-specific activation, or the DRM methyltransferase for locus-specific DNA-methylation. Although this method can increase the target gene’s transcript levels, it is not always enough to trigger a significant phenotype. In the same way, targeted DNA-methylation also initially presented some limitations of specificity and inheritability. However, the authors provide ways to overcome these problems and achieve desirable outputs (e.g., more gRNA, or by altering SunTag linker length, or effector nuclear localization signals). This paper is a good example of how a thorough analysis within a design-build-test-learn cyclic framework helps to the understanding of a technology and the observation of interesting phenomena. CRISPR-Cas9-SunTag system is a versatile expansion to the plant CRISPR toolbox. (Summary by Facundo Romani) Nature Comms. 10.1038/s41467-019-08736-7