Single cell multiomic analysis of plant immunity reveals PRIMER cells
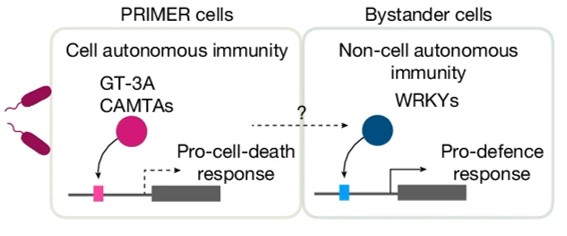
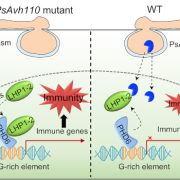
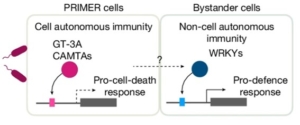 Single cell mutiomics are radically changing our understanding of pretty much every cellular process. Here, Nobori et al. integrated single-cell transcriptomic, epigenomic and spatial transcriptomic data to investigate plant responses to pathogens. The authors used three different strains of Pseudomonas syringae pv. tomato DC3000; the wild-type (referred to as DC3000) which causes disease, and two avirulent variants, DC3000 AvrRpt2 and AvrRpm1 that produce effectors that trigger the plant immune response. Nuclei were isolated at various times after inoculation for single-nucleus RNA-seq and Single-nucleus assay for transposase-accessible chromatin followed by sequencing (snATAC–seq), a method for identifying chromosome regions accessible to transcription factors and likely to indicate active genes. These large datasets revealed a wealth of information, including the fact that a subset of cellsact as primary immune responder (PRIMER) cells. These PRIMER cells express a novel transcription factor and are surrounded by “bystander” cells that activate systemic responses. The datasets are publicly available at (https://plantpathogenatlas.salk.edu). (Summary by Mary Williams) Nature https://doi.org/10.1038/s41586-024-08383-z
Single cell mutiomics are radically changing our understanding of pretty much every cellular process. Here, Nobori et al. integrated single-cell transcriptomic, epigenomic and spatial transcriptomic data to investigate plant responses to pathogens. The authors used three different strains of Pseudomonas syringae pv. tomato DC3000; the wild-type (referred to as DC3000) which causes disease, and two avirulent variants, DC3000 AvrRpt2 and AvrRpm1 that produce effectors that trigger the plant immune response. Nuclei were isolated at various times after inoculation for single-nucleus RNA-seq and Single-nucleus assay for transposase-accessible chromatin followed by sequencing (snATAC–seq), a method for identifying chromosome regions accessible to transcription factors and likely to indicate active genes. These large datasets revealed a wealth of information, including the fact that a subset of cellsact as primary immune responder (PRIMER) cells. These PRIMER cells express a novel transcription factor and are surrounded by “bystander” cells that activate systemic responses. The datasets are publicly available at (https://plantpathogenatlas.salk.edu). (Summary by Mary Williams) Nature https://doi.org/10.1038/s41586-024-08383-z