Security Notice: This Plant Immunity is under mRNA Surveillance
Every manufacture relies on a quality control process to ensure that the released product is without defects. Similarly, each transcript leaving the eukaryotic nucleus is subjected to mRNA surveillance, which helps to ensure that only flawless mRNAs are directed to translation. For example, transcripts in which the termination codon occurs upstream of the splice junction do not “make sense” for a cell, since translating them would lead to the production of truncated proteins. Instead, such transcripts are recognized and then destroyed via a process called Nonsense-Mediated Decay (NMD).
NMD also controls the abundance of normal transcripts, usually triggered by environmental cues. In this case NMD substrates are marked by a long (>300 nucleotides) 3’ untranslated region (UTR), an intron in the 3’ UTR, or an upstream open reading frame (ORF) which is out of frame with the protein coding sequence. Recognition of NMD-sensitive features and directing “marked” transcripts for decay requires a multi‑protein surveillance complex, with UPF (UP-FRAMESHIFT) proteins as its core members (Shaul, 2015). Mutants with impaired NMD accumulate transcripts related to biotic stress responses and have enhanced pathogen resistance (Gloggnitzer et al., 2014).
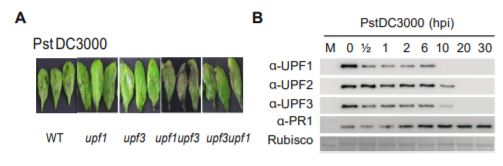
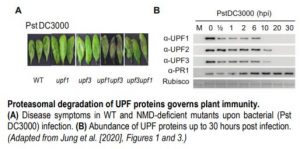 In recent work, Jung et al. (2020) shed light on the mechanism by which NMD shapes plant defense responses. The authors took a closer look at two members of the NMD complex, UPF1 and UPF3, which participate in NMD target recognition. Upon bacterial infection, the double mutant upf3 upf1 developed severe necrosis indicating a boost in its autoimmune response (see figure). Concurrently, elevated levels of the majority of Resistance (R) transcripts carrying NMD-eliciting signatures were detected in the upf3 upf1 mutant. By blocking de novo transcription and assessing the rate of transcript degradation, the authors demonstrated that R transcripts were stabilized in the upf3 upf1 mutant, whereas in wild type plants they were subjected to decay. This means that most R genes may be under the control of NMD (Jung et al., 2020).
In recent work, Jung et al. (2020) shed light on the mechanism by which NMD shapes plant defense responses. The authors took a closer look at two members of the NMD complex, UPF1 and UPF3, which participate in NMD target recognition. Upon bacterial infection, the double mutant upf3 upf1 developed severe necrosis indicating a boost in its autoimmune response (see figure). Concurrently, elevated levels of the majority of Resistance (R) transcripts carrying NMD-eliciting signatures were detected in the upf3 upf1 mutant. By blocking de novo transcription and assessing the rate of transcript degradation, the authors demonstrated that R transcripts were stabilized in the upf3 upf1 mutant, whereas in wild type plants they were subjected to decay. This means that most R genes may be under the control of NMD (Jung et al., 2020).
Surprisingly, UPF1 and UPF3 transcripts were upregulated by exposure to the pathogen. However, bacterial inoculation caused a decrease in all three UPF protein levels, but not in other components of the NMD machinery. The dynamics of protein turnover differed between individual UPF proteins, yet within 20 hours after infection, none of them were detectable (see figure). UPF protein decay was later linked to their increased ubiquitination and subsequent proteasomal degradation. Not all of the R transcripts with NMD-inducing signatures exhibited the same level of susceptibility to NMD, which suggests that other post-transcriptional mechanisms might act in parallel to regulate their abundance. Further, overexpression of two of the R genes whose transcripts accumulated in upf3 upf1 mutant resulted in enhanced resistance to bacterial infection (Jung et al., 2020).
Upon detection of the presence of a pathogen, R protein activity turns on the defense mode. However, a prolonged immune response might eventually compromise growth and plant fitness (Karasov et al., 2017). Equipping R transcripts with NMD-inducing signatures enables fine tuning of R transcript abundance and restricts the defense response to times when it is most needed. In the model proposed by Jung et al. (2020), proteasomal degradation of UPF proteins disarms NMD machinery to protect a broad range of R transcripts and prepare a plant to fight against a variety of pathogens. Plant gain immunity, yet might still pay a price for becoming more vulnerable to other nonsense messages.
Dorota Kawa
Department of Plant Biology and Genome Center
University of California, Davis
ORCID: 0000-0002-4227-1621
REFERENCES
Gloggnitzer, J., Akimcheva, S., Srinivasan, A., Kusenda, B., Riehs, N., Stampfl, H., Bautor, J., Dekrout, B., Jonak, C., Jimenez-Gomez, J.M., Parker, J.E., and Riha, K. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16, 376-390.
Karasov, T.L., Chae, E., Herman, J.J., and Bergelson, J. (2017). Mechanisms to Mitigate the Trade-Off between Growth and Defense. Plant Cell 29, 666-680.
Shaul, O. (2015). Unique Aspects of Plant Nonsense-Mediated mRNA Decay. Trends Plant Sci 20, 767-779.
Jung, H.W., Panigrahi, G.K., Jung, G-Y., Lee, J., Shin, K.H., Sahoo, A., Choi, E.S., Lee, E., Kim, K.M., Yang, S.H, Jeon, J-S, Lee, S.C., Kim, S.H. (2020). PAMP-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during the early defense response. The Plant Cell. https://doi.org/10.1105/tpc.19.00631