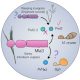
RNA composition of Processing Bodies
Liu et al. explore the RNAs present in processing bodies, a cytoplasmic subcellular condensate.
Andriani Mentzelopoulou
Department of Biology, University of Crete, Heraklion, Greece
Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology–Hellas, Heraklion, Greece
Panagiotis Moschou
Department of Biology, University of Crete, Heraklion, Greece
Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology–Hellas, Heraklion, Greece
Department of Molecular Sciences, Uppsala BioCenter, Swedish University of Agricultural Sciences and Linnean Center for Plant Biology, Uppsala, Sweden
https://doi.org/10.1093/plcell/koad288
Background: Interactions between proteins and nucleic acids can promote the generation of droplet-like ribonucleoprotein condensates. One example of a condensate is Processing Bodies (PBs), which are found in the cytoplasm and contain many proteins and RNA molecules. Previously, we defined the proteins residing in Arabidopsis thaliana PBs using an approach known as proximity biotinylation, which can capture weak and transient interactions, such as those occurring in condensates. We did that in two conditions, normal conditions, and heat stress, showing that PBs are highly dynamic. This approach allowed us to identify a link between the core PB component known as DECAPPING 1 (DCP1) and the SCAR/WAVE actin nucleating complex.
Question: Having defined the PB proteome, we asked whether the proximity biotinylation approach can also be used to capture associated RNA molecules and what kind of RNA molecules reside in PBs.
Findings: Herein, we used Arabidopsis thaliana seedlings and especially roots. We showed that the proximity biotinylation approach can indeed capture RNAs stably to more transiently associated with the PBs. These mainly include RNAs involved in cell wall development and regeneration, plant hormonal signaling, secondary metabolism/defense, and RNA metabolism. Furthermore, we showed that RNAs in PBs follow different paths: in small PBs, RNAs get degraded; in larger PBs, RNAs are stored. The stored RNAs are not translated until the actin-related SCAR/WAVE complex dissolves PBs and releases specific RNAs. One example of such RNA is WOUND INDUCED DEDIFFERENTIATION 1/RELATED TO AP2 4 (RAP2.4), which affects responses to the phytohormone ethylene.
Next steps: We could use the findings from this study to define PB functions in a cell-specific manner during development of the Arabidopsis root. The approaches developed here can also be used to decipher the composition of numerous other condensates.
Reference:
Chen Liu, Andriani Mentzelopoulou, Ioannis H. Hatzianestis, Epameinondas Tzagkarakis, Vassilis Scaltsoyiannes, Xuemin Ma, Vassiliki A. Michalopoulou, Francisco J. Romero–Campero, Ana B. Romero–Losada, Panagiotis F. Sarris, Peter Marhavy, Bettina Bölter, Alexandros Kanterakis1, Emilio Gutierrez–Beltran, Panagiotis N. Moschou (2023) A proxitome-RNA-capture approach reveals that processing bodies repress co-regulated hub genes https://doi.org/10.1093/plcell/koad288
mRNA处理小体的RNA构成
蛋白质和核酸之间的相互作用促进了液滴状核糖核蛋白(RNP)凝聚体的形成。mRNA处理小体(PBs)就是细胞质中的一类凝聚物。这些mRNA处理小体中富含蛋白质和 RNA 分子。我们在之前的研究中曾使用一种能捕捉微弱且短暂的蛋白相互作用(如在凝聚体中发生的相互作用)的方法(即邻近生物素标记技术)来解析富集在 mRNA处理小体中的蛋白质组。我们比较了正常条件和热胁迫条件下的RNA-seq数据,结果表明 mRNA处理小体具有高度动态性。通过这种方法,我们确定了 mRNA处理小体的核心部件脱帽蛋白1-DECAPPING 1 (DCP1) 与 SCAR/WAVE 肌动蛋白成核复合体之间的紧密联系。
在解析了 PBs 的邻近蛋白质组之后,我们尝试研究邻近标记技术是否也能用于捕获与之关联的 RNA 分子,以及哪些类型的 RNA 分子聚集于mRNA处理小体中。
在本文中,我们以拟南芥幼苗(尤其是它的根)作为材料,揭示了邻近标记技术确实可以捕获与PBs稳定或瞬时相关的RNAs。其中主要包括参与细胞壁发育和再生、植物激素信号转导、次生代谢/防御以及RNA代谢的RNAs。实验还发现,mRNA处理小体PBs 中的 RNAs有不同的命运:在小的PBs中,RNAs 通常会被降解,而在大的PBs中,RNAs 会被储存起来。储存的 RNA不会被翻译,直到肌动蛋白相关复合物 SCAR/WAVE 促使PBs解体并释放出特定的 RNA。伤口诱导去分化 1(WOUND INDUCED DEDIFFERENTIATION 1)/AP2.4相关蛋白(RELATED TO AP2 4(RAP2.4))就是这类 RNA的一个例子,它影响植物对乙烯的应答。
该研究结果可用于以细胞特异性的方式沿着发育的拟南芥根轴来确定mRNA处理小体PBs 的功能。而该研究提出的方法可用于破译许多其他类似凝聚体的构成。
凝聚体、液液相分离、邻近标记技术、乙烯