Barley MLA3 recognizes the blast effector Pwl2, an effector involved in host range dynamics
Brabham et al. explore the mechanisms of resistance to fungal pathogens in barley.
Helen J. Brabham1,2, Diana Gómez De La Cruz1, Matthew J. Moscou,3
1The Sainsbury Laboratory, University of East Anglia, Norwich Research Park, Norwich, NR4 7UH, UK
22Blades, 1630 Chicago Avenue, Suite 1312, Evanston IL 60201, USA
3USDA-ARS, Cereal Disease Laboratory, St. Paul, MN 55108, USA
https://doi.org/10.1093/plcell/koad266
Background: Nucleotide-binding leucine-rich repeat (NLR) proteins are intracellular immune receptors that recognize molecules from plant pathogens, known as effectors. The majority of NLRs confer resistance to one pathogen by recognizing a specific effector. In barley (Hordeum vulgare), the Mla locus contains multiple NLR genes and alleles of the NLR Mla recognize Blumeria graminis, the causal agent of powdery mildew. In the barley cultivar Baronesse, resistance to the blast pathogen Magnaporthe oryzae has been mapped to the Mla locus, but the causal gene within this locus is unknown.
Question: How is the barley cultivar Baronesse resistant to multiple pathogens? Which gene in the Mla locus provides resistance to the rice blast pathogen? Does the Mla allele Mla3 in Baronesse recognize M. oryzae in addition to B. graminis? What effector is being recognized from M. oryzae?
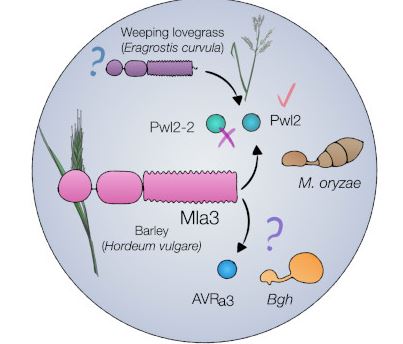 Findings: We show that the barley NLR MLA3 recognizes the effector Pwl2 from M. oryzae. Resistance to blast disease was mapped to the Mla locus. Three candidate genes were cloned and introduced into a susceptible barley cultivar. Infection assays with M. oryzae in transgenic barley lines found that only barley carrying Mla3 showed resistance and required multiple copies. To identify the M. oryzae effector recognized by Mla3, mutants were generated by randomly knocking genes out in M. oryzae using UV mutagenesis. Sequencing mutants found that the effector gene PWL2 was always lost. PWL2 was first discovered in 1995, as it prevents blast isolates from infecting weeping lovegrass (Eragrostis curvula). MLA3 was shown to directly recognize and associate with Pwl2 through expression and protein-protein assays.
Findings: We show that the barley NLR MLA3 recognizes the effector Pwl2 from M. oryzae. Resistance to blast disease was mapped to the Mla locus. Three candidate genes were cloned and introduced into a susceptible barley cultivar. Infection assays with M. oryzae in transgenic barley lines found that only barley carrying Mla3 showed resistance and required multiple copies. To identify the M. oryzae effector recognized by Mla3, mutants were generated by randomly knocking genes out in M. oryzae using UV mutagenesis. Sequencing mutants found that the effector gene PWL2 was always lost. PWL2 was first discovered in 1995, as it prevents blast isolates from infecting weeping lovegrass (Eragrostis curvula). MLA3 was shown to directly recognize and associate with Pwl2 through expression and protein-protein assays.
Next steps: The main question that remains to be addressed is how does MLA3 recognize M. oryzae and B. graminis? Future efforts will involve finding the molecular principles of Pwl2 recognition by MLA3 and identifying the recognized effector of B. graminis to understand the mechanism by which MLA3 recognizes multiple pathogens.
Reference:
Helen J. Brabham, Diana Gómez De La Cruz, Vincent Were, Motoki Shimizu, Hiromasa Saitoh, Inmaculada Hernández-Pinzón, Phon Green, Jennifer Lorang, Koki Fujisaki, Kazuhiro Sato, István Molnár, Hana Šimková, Jaroslav Doležel, James Russell, Jodie Taylor, Matthew Smoker, Yogesh Kumar Gupta, Tom Wolpert, Nicholas J. Talbot, Ryohei Terauchi, Matthew J. Moscou (2023) Barley MLA3 recognizes the host-specificity effector Pwl2 from Magnaporthe oryzae. https://doi.org/10.1093/plcell/koad266