Review: Sequencing and analyzing the transcriptomes of a thousand plant species (Annu. Rev. Plant Biol.)
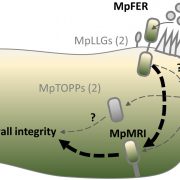
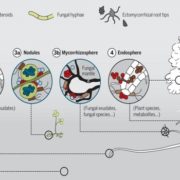
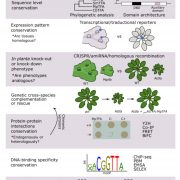
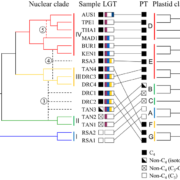
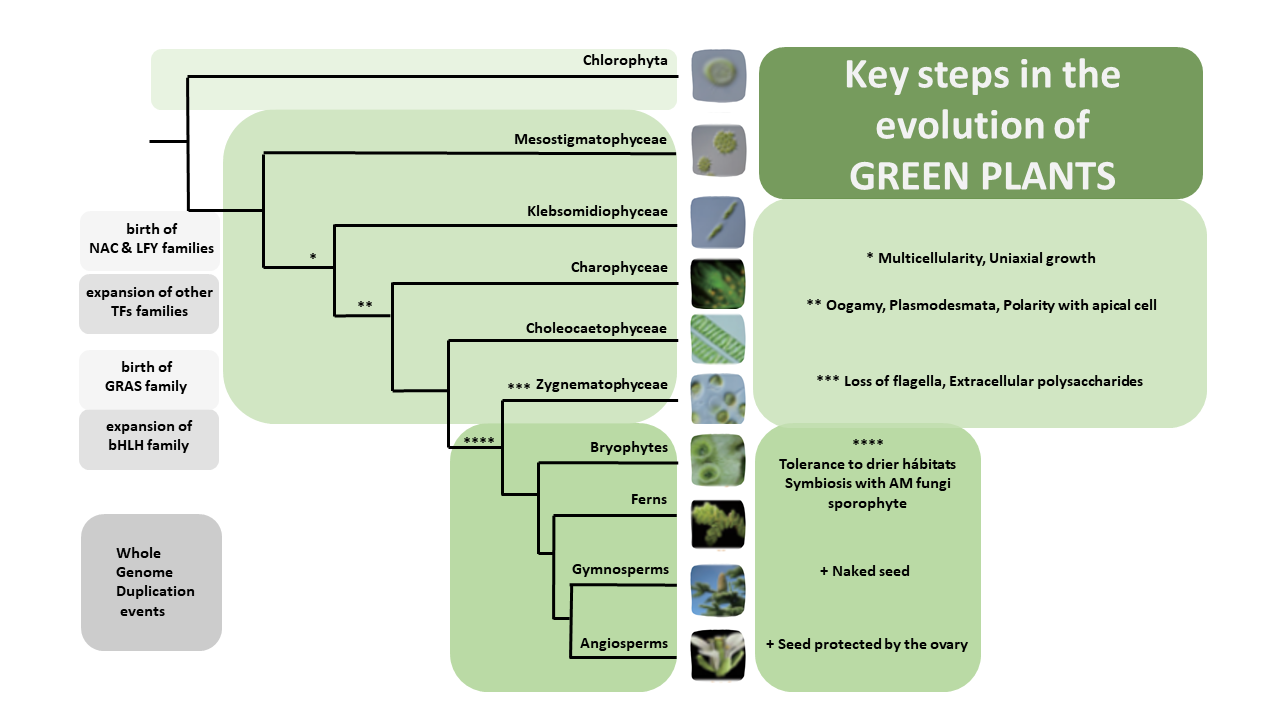 Over the past decade, Next Generation Sequencing (NGS) has been used for de novo assembly of crop genomes (i.e., tomato, potato) under the motto “If it tastes good, let’s sequence it”. By contrast, the One Thousand Plant (1KP) Initiative set out to obtain transcriptomic data of phylogenetically diverse green plant species without any apparent economic importance. Wong et al. review how the biodiversity in this large dataset is a valuable tool not only to address evolutionary questions but also to find original resources for genetic engineering. Phylotranscriptomic analysis of the 1KP dataset sheds new light on the diversification of green plant lineages, uncovering 138 novel Whole-Genome Duplication (WGD) events. Interestingly, paleopolyploidy was not found in the lycophyte Selaginella whereas at least five rounds of WGDs occurred in many angiosperms. Novel insights were gained on the origin and expansion of important multigene families encoding regulatory proteins. Surprisingly, regulatory genes thought to be specific to angiosperms, such as genetic determinants of flower development, were already present early in the evolution of streptophytes, the closest algal relatives of land plants. The 1KP dataset was also explored for biotechnological applications including studies of light-sensitive proteins to be employed in neuroscience and the elucidation of genetic networks underlying complex traits such as C4 photosynthesis. It will soon be feasible to sequence up to 10,000 genomes due to decreasing costs of NGS; these additional data will clarify discordances for critical nodes in the plant phylogenetic tree, and likely solve mysteries about the origin and evolution of green plants. (Summary and graphical abstract by Michela Osnato @michela_osnato) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042916-041040
Over the past decade, Next Generation Sequencing (NGS) has been used for de novo assembly of crop genomes (i.e., tomato, potato) under the motto “If it tastes good, let’s sequence it”. By contrast, the One Thousand Plant (1KP) Initiative set out to obtain transcriptomic data of phylogenetically diverse green plant species without any apparent economic importance. Wong et al. review how the biodiversity in this large dataset is a valuable tool not only to address evolutionary questions but also to find original resources for genetic engineering. Phylotranscriptomic analysis of the 1KP dataset sheds new light on the diversification of green plant lineages, uncovering 138 novel Whole-Genome Duplication (WGD) events. Interestingly, paleopolyploidy was not found in the lycophyte Selaginella whereas at least five rounds of WGDs occurred in many angiosperms. Novel insights were gained on the origin and expansion of important multigene families encoding regulatory proteins. Surprisingly, regulatory genes thought to be specific to angiosperms, such as genetic determinants of flower development, were already present early in the evolution of streptophytes, the closest algal relatives of land plants. The 1KP dataset was also explored for biotechnological applications including studies of light-sensitive proteins to be employed in neuroscience and the elucidation of genetic networks underlying complex traits such as C4 photosynthesis. It will soon be feasible to sequence up to 10,000 genomes due to decreasing costs of NGS; these additional data will clarify discordances for critical nodes in the plant phylogenetic tree, and likely solve mysteries about the origin and evolution of green plants. (Summary and graphical abstract by Michela Osnato @michela_osnato) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042916-041040