Review: Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding (Nature Genetics)
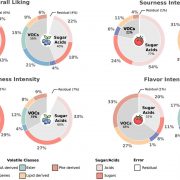
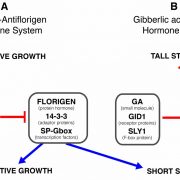
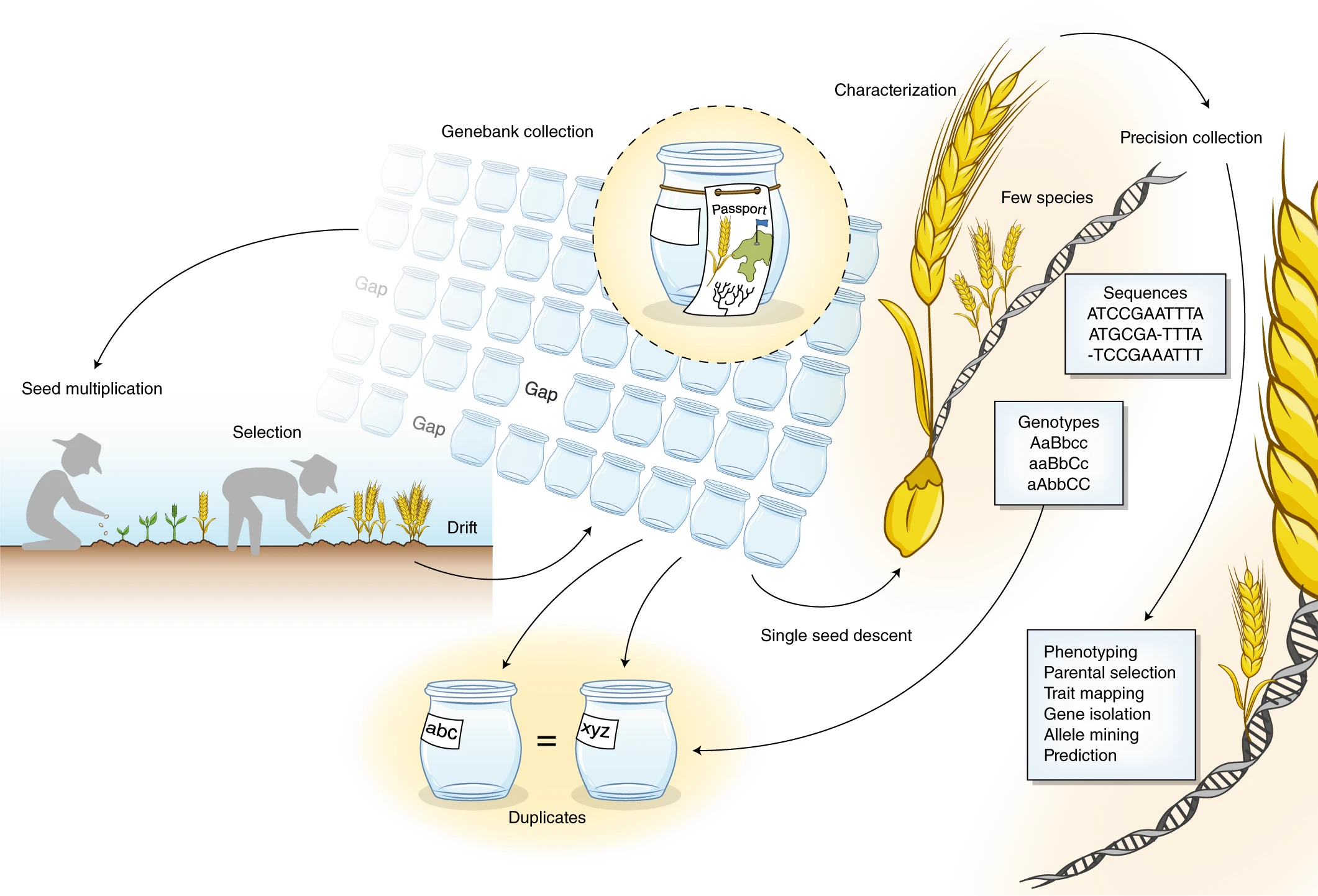 Crop diversity is fundamental to safeguarding global food security. The high-yielding, input-responsive cultivars developed post-green revolution led to the replacement of traditional landraces that harbour beneficial genes and alleles governing biotic and abiotic stress resistance and nutritional quality improvement. Genebanks conserve these invaluable plant genetic resources (PGRs) for future crop improvement. The review by Masher et al. is a beautiful narration of the author’s vision for the transformation of genebanks into bio-digital resource centres through application of genomic approaches in genebank management and pre-breeding activities. Molecular data on single nucleotide polymorphisms obtained through high-throughput genotyping platforms can complement the conventional passport records in defining the identity of the collections. Duplicate accessions that escalate the cost and workload to genebank managers can be cleansed through identity-by-state analyses using marker data from large number of individuals. As phenotyping the entire collection for complex traits like yield and quality is burdensome, initial core sets can be created based on maximized neutral genetic diversity using the marker data. The phenotypes of the remaining accessions can be imputed from the prediction models developed using the phenotypes of the core sets coupled with marker data. Genome wide association mapping using such core sets would directly lead to the identification of the candidate gene. This can be followed by allele mining to determine allelic diversity in the collections. The promising genotypes thus identified with superior alleles can be crossed to modern breeding lines, and speed breeding technologies could hasten the development of commercial varieties. (Summary by Haritha Bollinedi) Nature Genetics 10.1038/s41588-019-0443-6
Crop diversity is fundamental to safeguarding global food security. The high-yielding, input-responsive cultivars developed post-green revolution led to the replacement of traditional landraces that harbour beneficial genes and alleles governing biotic and abiotic stress resistance and nutritional quality improvement. Genebanks conserve these invaluable plant genetic resources (PGRs) for future crop improvement. The review by Masher et al. is a beautiful narration of the author’s vision for the transformation of genebanks into bio-digital resource centres through application of genomic approaches in genebank management and pre-breeding activities. Molecular data on single nucleotide polymorphisms obtained through high-throughput genotyping platforms can complement the conventional passport records in defining the identity of the collections. Duplicate accessions that escalate the cost and workload to genebank managers can be cleansed through identity-by-state analyses using marker data from large number of individuals. As phenotyping the entire collection for complex traits like yield and quality is burdensome, initial core sets can be created based on maximized neutral genetic diversity using the marker data. The phenotypes of the remaining accessions can be imputed from the prediction models developed using the phenotypes of the core sets coupled with marker data. Genome wide association mapping using such core sets would directly lead to the identification of the candidate gene. This can be followed by allele mining to determine allelic diversity in the collections. The promising genotypes thus identified with superior alleles can be crossed to modern breeding lines, and speed breeding technologies could hasten the development of commercial varieties. (Summary by Haritha Bollinedi) Nature Genetics 10.1038/s41588-019-0443-6