Review: Deep learning approaches to understanding stomatal function
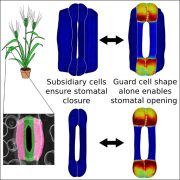
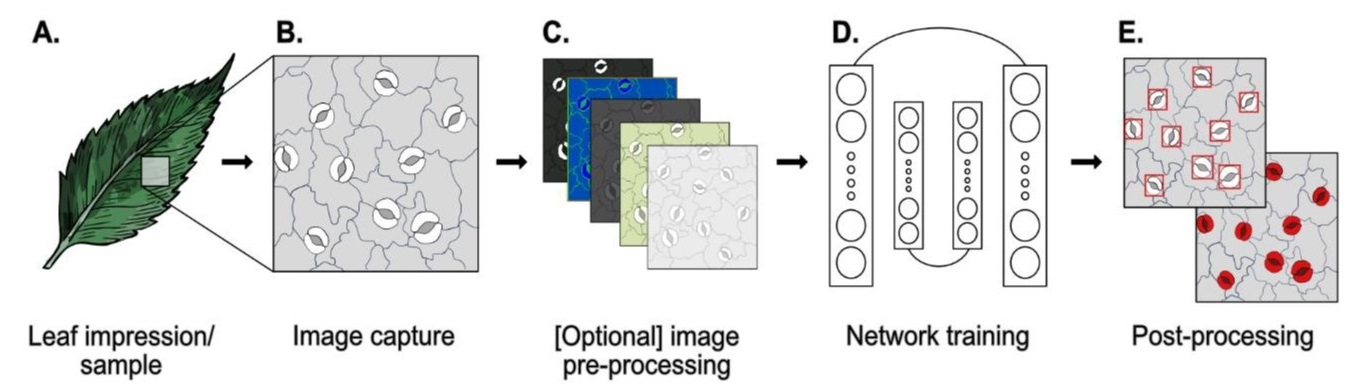 Sydney Brenner famously said, “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Right now, we’re seeing how advancements and new techniques in artificial intelligence and deep learning are being applied in plant sciences, including in the analysis of all sorts of large -omics datasets. This review by Gibbs and Burgess discusses deep learning approaches to understand stomatal function, and it provides a good overview of what is inside the (to me) black box of deep learning. Stomatal function depends on stomatal morphology, largely determined during development, and the very dynamic stomatal aperture, which responds to environment and physiology. The authors explain the steps in acquiring data, building models, and testing the models, and summarize methods and limitations for the study of stomata. They call for greater communication amongst those developing deep learning pipelines and greater transparency and sharing of datasets, and to encourage such collaboration and sharing they have created StomataHub (http://www.stomatahub.com/). Finally, they note that knowledge is currently poorly translated to field studies, which are integral for yield improvement, but that such translation is needed to generate new discoveries and ideas. (Summary by Mary Williams @PlantTeaching) J Exp Bot 10.1093/jxb/erae207
Sydney Brenner famously said, “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Right now, we’re seeing how advancements and new techniques in artificial intelligence and deep learning are being applied in plant sciences, including in the analysis of all sorts of large -omics datasets. This review by Gibbs and Burgess discusses deep learning approaches to understand stomatal function, and it provides a good overview of what is inside the (to me) black box of deep learning. Stomatal function depends on stomatal morphology, largely determined during development, and the very dynamic stomatal aperture, which responds to environment and physiology. The authors explain the steps in acquiring data, building models, and testing the models, and summarize methods and limitations for the study of stomata. They call for greater communication amongst those developing deep learning pipelines and greater transparency and sharing of datasets, and to encourage such collaboration and sharing they have created StomataHub (http://www.stomatahub.com/). Finally, they note that knowledge is currently poorly translated to field studies, which are integral for yield improvement, but that such translation is needed to generate new discoveries and ideas. (Summary by Mary Williams @PlantTeaching) J Exp Bot 10.1093/jxb/erae207