Small RNA Pathway Acting in the Father Controls Seed Abortion
Satyaki and Mary Gehring uncover the mechanism underlying interploidy seed abortion, challenging previous models of dosage sensitivity in endosperm. Plant Cell https://doi.org/10.1105/tpc.19.00047
By PRV Satyaki (Whitehead Institute for Biomedical Research) and Mary Gehring (Whitehead Institute for Biomedical Research and Department of Biology, Massachusetts Institute of Technology)
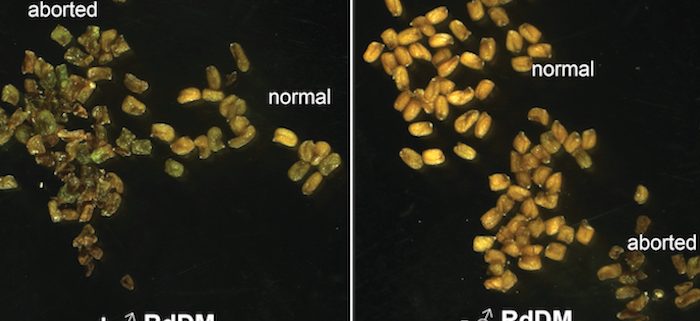
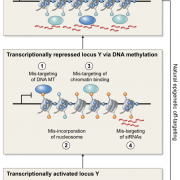
Background: In the seeds of flowering plants, the embryo is surrounded by a tissue called the endosperm, which mediates nutrient transfer between the mother and the growing embryo. Unlike the rest of the plant, the endosperm contains two copies of the mother’s genome and one copy of the father’s. This unusual ratio of maternal-to-paternal genomes is important for normal endosperm development and seed viability. Seeds with extra genomes from the father abort due to defective endosperm development (interploidy seed abortion). Defective development is hypothesized to be due to mis-regulation of either imprinted genes (genes expressed primarily from one parent’s copy) or transposons. We previously showed that the epigenetic regulator RNA Pol IV is essential for Arabidopsis interploidy seed abortion. Clarifying the cause of seed abortion and the role of RNA Pol IV is important for understanding seed development.
Question: How does the lack of RNA Pol IV prevent interploidy seed abortion? Where does RNA Pol IV act–in the endosperm or in the father–to influence gene expression in the endosperm? What genetic pathway does RNA Pol IV function in to cause seed abortion?
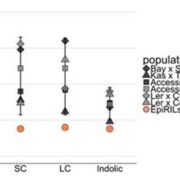
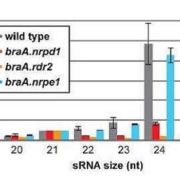
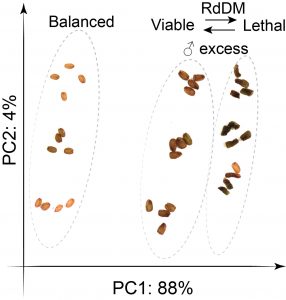 Findings: RNA Pol IV acts in the father along with other genes in the canonical RNA-directed DNA methylation pathway (which uses small RNAs to guide DNA methylation) to cause seed abortion. We compared endosperm transcriptomes of interploidy seeds that aborted to those that were viable due to the loss of paternal RNA Pol IV. Remarkably, transposons and thousands of genes (including imprinted genes) were mis-regulated in both living and dying seeds. Transposons and imprinted genes, therefore, do not appear to drive seed abortion. Rather, mis-regulation of relatively few genes distinguished living from aborting seeds. This work also revealed a transcriptional buffering system in the endosperm that counteracts increased paternal genome dose by reducing the expression of the paternal copy of some genes and increasing the expression of the maternal copy of others.
Findings: RNA Pol IV acts in the father along with other genes in the canonical RNA-directed DNA methylation pathway (which uses small RNAs to guide DNA methylation) to cause seed abortion. We compared endosperm transcriptomes of interploidy seeds that aborted to those that were viable due to the loss of paternal RNA Pol IV. Remarkably, transposons and thousands of genes (including imprinted genes) were mis-regulated in both living and dying seeds. Transposons and imprinted genes, therefore, do not appear to drive seed abortion. Rather, mis-regulation of relatively few genes distinguished living from aborting seeds. This work also revealed a transcriptional buffering system in the endosperm that counteracts increased paternal genome dose by reducing the expression of the paternal copy of some genes and increasing the expression of the maternal copy of others.
Next steps: We plan to unravel the mechanism underlying the transcriptional buffering system and to identify the genes responsible for interploidy seed abortion using the shortlist of candidate genes generated from our transcriptional studies.
P.R.V. Satyaki and Mary Gehring (2019). Paternally Acting Canonical RNA-directed DNA Methylation Pathway Genes Sensitize Arabidopsis Endosperm to Paternal Genome Dosage. Plant Cell 31: xx; https://doi.org/10.1105/tpc.19.00047
Key words: Epigenetics, parental effect, seed development, interploidy, RdDM