Regulation of Imprinting in Arabidopsis
The seeds of flowering plants consist of three genetically distinct components: the diploid embryo with one genome copy from each of the parents, the triploid endosperm with two maternal copies and one paternal copy, and the seed coat having the same genotype as the diploid mother plant. Balanced parental gene expression for proper development of progeny is important, and the endosperm in particular is a site of genomic imprinting, an epigenetic phenomenon that leads to parent-of-origin-dependent expression of genes due to non-DNA sequence-based mechanisms established in the male and female germline. Parent-of-origin-dependent expression is independent of the underlying DNA sequence and relies on epigenetic 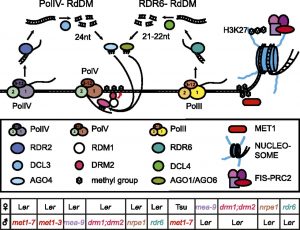 mechanisms that include DNA methylation and modifications of histones. To investigate imprinting more fully, Hornslien et al. (10.1104/pp.19.00320) have used high-resolution sequencing data to examine parent-of-origin allele-specific expression. They report the occurrence of more than 300 loci with parental expression bias in Arabidopsis. In most cases, the level of expression from maternal and paternal alleles was not binary, instead supporting a differential dosage hypothesis for the evolution of imprinting in plants. To gain further insights into imprinting regulation, they systematically employed mutations in regulatory epigenetic pathways proposed to be major players in the process, and thereby establish that imprinting for more than 50 loci is regulated by DNA methylation and Polycomb-dependent histone methylation. However, the imprinting patterns of most genes were not affected by these mechanisms. They also demonstrate that the RNA-directed DNA methylation pathway alone does not substantially influence imprinting patterns, suggesting that unknown or more complex epigenetic pathways regulate the majority of identified imprinted genes.
mechanisms that include DNA methylation and modifications of histones. To investigate imprinting more fully, Hornslien et al. (10.1104/pp.19.00320) have used high-resolution sequencing data to examine parent-of-origin allele-specific expression. They report the occurrence of more than 300 loci with parental expression bias in Arabidopsis. In most cases, the level of expression from maternal and paternal alleles was not binary, instead supporting a differential dosage hypothesis for the evolution of imprinting in plants. To gain further insights into imprinting regulation, they systematically employed mutations in regulatory epigenetic pathways proposed to be major players in the process, and thereby establish that imprinting for more than 50 loci is regulated by DNA methylation and Polycomb-dependent histone methylation. However, the imprinting patterns of most genes were not affected by these mechanisms. They also demonstrate that the RNA-directed DNA methylation pathway alone does not substantially influence imprinting patterns, suggesting that unknown or more complex epigenetic pathways regulate the majority of identified imprinted genes.



