RDR6 is essential for double-strand break formation during male meiosis in rice
Plant RNA-dependent RNA polymerases (RDRs) are essential for the biogenesis of small, interfering RNAs (siRNAs). These polymerases function by converting single-stranded RNA transcripts into double-stranded RNAs, which are processed by Dicer-like ribonucleases into 21- to 24-nucleotide siRNAs (reviewed in Borges and Martienssen, 2015). RDR6 in rice (Oryza sativa) has been shown to be required for shoot apical meristem and floral organ development (Nagasaki et al., 2007; Song et al., 2012). However, its role in meiosis remains uncharacterized.
Liu et al. (2020) identified a rice mutant that is male and female sterile, producing no viable pollen or functional embryo sac. The phenotype is recessive, and is controlled by a single nuclear gene. The authors showed that the underlying mutation is a missense point mutation in the OsRDR6 gene, and this new Osrdr6 allele was named Osrdr6-mei based on the defects in meiosis. A genetic complementation analysis verified OsRDR6 as the causal gene of the reproductive defects.
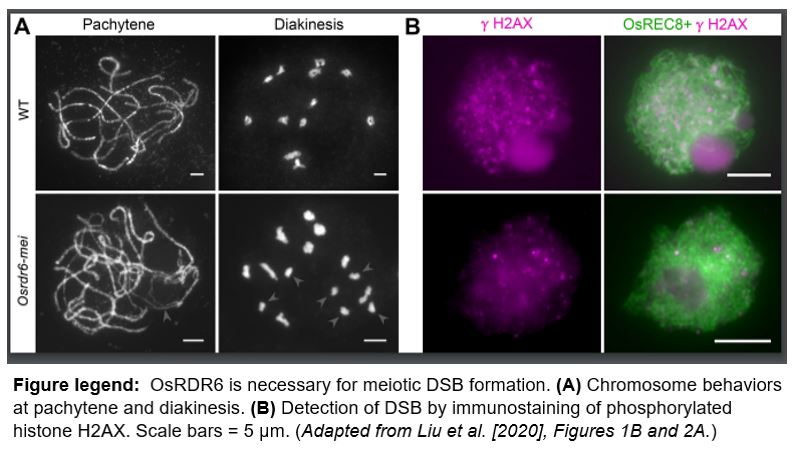 To understand the role of OsRDR6 in meiotic progression, the authors examined chromosome behavior during male meiosis in the Osrdr6-mei mutant. In wild type, all homologous chromosomes in meiocytes are paired at prophase I; whereas in the mutant, some homologous chromosomes are not paired. Furthermore, the authors showed that the formation of DNA double-strand breaks (DSBs), which are crucial for crossover formation during meiosis, is perturbed in the Osrdr6-mei mutant (see figure).
To understand the role of OsRDR6 in meiotic progression, the authors examined chromosome behavior during male meiosis in the Osrdr6-mei mutant. In wild type, all homologous chromosomes in meiocytes are paired at prophase I; whereas in the mutant, some homologous chromosomes are not paired. Furthermore, the authors showed that the formation of DNA double-strand breaks (DSBs), which are crucial for crossover formation during meiosis, is perturbed in the Osrdr6-mei mutant (see figure).
RNA sequencing detected hundreds of differentially expressed genes between wild-type and Osrdr6-mei anthers. Among the downregulated genes, three genes (OsSDS, P31comet, and CRC1) have previously been shown to be essential for meiotic DSB formation in rice. The authors speculated that OsRDR6 regulates meiotic DSB formation by modulating the expression levels of these genes.
The authors further showed that the abundance of 21-nucleotide small RNAs (21-mers) is dramatically downregulated in the mutant, whereas the abundance of 24-mers is upregulated, suggesting that small RNA biogenesis or turnover processes are dysregulated in the mutant. 24-nucleotide siRNAs in plants are known to direct DNA methylation. Whole-genome bisulfite sequencing showed upregulation of methylation levels at transposable element loci and regions surrounding non-transposable element genes in the mutant anthers, suggesting that OsRDR6 plays a repressive role in the regulation of DNA methylation.
Based on the collective data, the authors proposed a model whereby the Osrdr6-mei mutant accumulates excessive 24-nucleotide small RNAs, which mediate silencing of genes involved in DSB formation. Since RDR6 is known as a canonical biogenesis factor of multiple types of small RNAs in plants (Borges and Martienssen, 2015), the challenge now is to identify the specific small RNAs, if any, that regulate meiotic DSB formation, and understand how they function during meiosis.
Junpeng Zhan
Donald Danforth Plant Science Center
St. Louis, Missouri
Department of Biology and Institute of Plant and Food Science
Southern University of Science and Technology
Shenzhen, Guangdong, China
[email protected]
ORCID ID: 0000-0001-7353-7608
REFERENCES
Borges, F., and Martienssen, R.A. (2015). The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16, 727-741.
Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., Mukouhata, M., Ashikari, M., Kitano, H., Matsuoka, M., Nagato, Y., and Sato, Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences of the United States of America 104, 14867-14871.
Song, X., Wang, D., Ma, L., Chen, Z., Li, P., Cui, X., Liu, C., Cao, S., Chu, C., Tao, Y., and Cao, X. (2012). Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J 71, 378-389.