Proximity labeling to examine viral replication complexes
Q. Zhang, Z. Wen, and X. Zhang, et al. use proximity labeling with the TurboID system to identify proteins that are important for replication in plant viruses.
https://doi.org/10.1093/plcell/koad146
Yongliang Zhang1, Xiaofeng Wang2
1 State Key Laboratory of Plant Environmental Resilience and Ministry of Agriculture Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China
2 School of Plant and Environmental Sciences, Virginia Tech, Blacksburg, Virginia 24061, USA
Background:
Upon entry into host cells, positive-strand RNA viruses hijack host factors to remodel specific organelle membranes to form viral replication complexes (VRCs). VRCs provide an optimal microenvironment for protecting and generating progeny viral RNAs. Identifying host components of VRCs is crucial for understanding viral replication mechanisms and developing virus-resistant plants. However, VRCs are membrane associated and dynamic, making it technically challenging to study VRC components using traditional methods for examining protein–protein interactions in plant cells. The recently emerged TurboID-based proximity labeling (PL) approach has become a powerful tool for examining weak, transient, and dynamic molecular interactions in planta. However, TurboID-based PL has not been employed to investigate VRCs in plants.
Question:
Can TurboID-based PL be used to identify key components and interaction networks of VRCs in plants?
Findings:
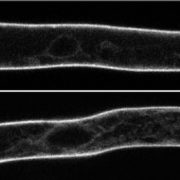
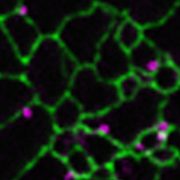
Using Beet black scorch virus (BBSV) as a model, we systematically investigated the constituents of VRCs by fusing the TurboID enzyme to the BBSV replication protein p23. We identified 185 p23-proximal proteins, including several proteins that are known to interact with p23 or to be critical for BBSV replication. The reticulon family proteins were repeatedly identified. We further demonstrated that reticulon-like protein B2 (RTNLB2) interacts with p23 and plays a pro-viral role in BBSV replication by facilitating the establishment of VRCs. Moreover, RTNLB2 is crucial for replication of several other plant viruses that replicate on endoplasmic reticulum membranes.
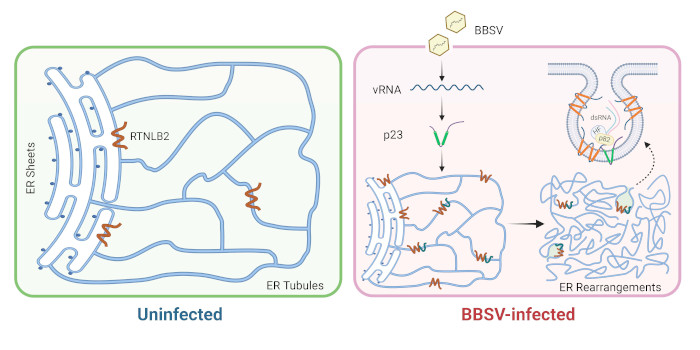 Next steps:
Next steps:
Whether RTNLB2 is involved in other aspects of virus infection, such as movement, remains to be investigated. Are other p23-proximal proteins involved in replications of BBSV and/or other plant viruses, if so, what are the underlying mechanisms?
Reference:
Qianshen Zhang, Zhiyan Wen, Xin Zhang, Jiajie She, Xiaoling Wang, Zongyu Gao, Ruiqi Wang, Xiaofei Zhao, Zhen Su, Zhen Li, Dawei Li, Xiaofeng Wang, Yongliang Zhang (2023). RETICULON-LIKE PROTEIN B2 is a pro-viral factor coopted for the biogenesis of viral replication organelles in plants. https://doi.org/10.1093/plcell/koad146