Proteomics-based protein complex discovery in the developing rice aleurone-subaleurone
Background: Many proteins act as components of multiprotein complexes that coordinate cellular processes such as vesicle trafficking, cell signaling, and metabolism. Moreover, the protein-protein interaction network functions as a dynamic system that can rearrange to enable productive responses to developmental or environmental changes. However, at present, knowledge about interaction networks is sparse.
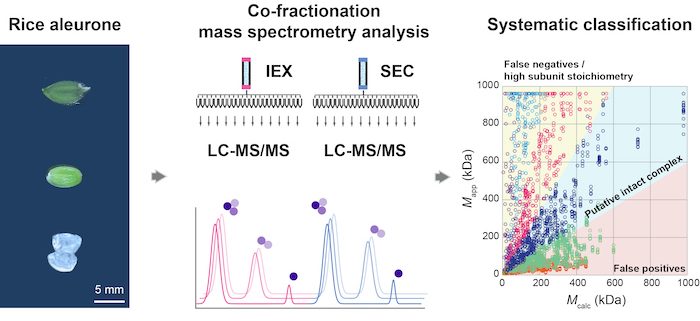
Question: We set out to determine if co-fractionation mass spectrometry (CFMS) can be used to generate reliable predictions about protein complex compositions in developing aleurone-subaleurone layers of rice (Oryza sativa) seeds.
Findings: The CFMS pipeline utilizes biological replicates, reproducibility filters, and orthogonal separations of soluble extracts to reduce noise and the confounding effect of chance co-elution. Based on a distance-based clustering and a validated classification system, the compositions of 770 reliable rice protein complex predictions were generated. The predictions included both known complexes/subcomplexes with diverse functions and many new ones. This snapshot of protein multimerization provides a systems-level view into the development and physiology of tissues crucial for rice seed quality.
Next steps: This approach and the associated data have significant value for the research community to broadly test hypotheses about protein complex function across species and cell types. This open-ended proteomics pipeline is being further developed to analyze the compositions of membrane-associated complexes and those that change in response to mutation and environmental stress.
Youngwoo Lee, Thomas W. Okita, and Daniel B. Szymanski (2021). A co-fractionation mass spectrometry-based prediction of protein complex assemblies in the developing rice aleurone-subaleurone. Plant Cell https://doi.org/10.1093/plcell/koab182