Plant Tomosyn is a negative regulator of SNARE-mediated secretion in pollen
Emily R Larson, [email protected]
Vesicle transport is one of those molecular processes that is easy to take for granted. From cell expansion and polarised growth to protein regulation and signalling, vesicle trafficking plays its unsung part. We tend to believe that we know a lot about this process and in truth, we do. Much work has been done to describe the players, particularly the SNAREs and their Rab GTPase drivers (Cai et al., 2007; Sanderfoot et al., 2000). SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) are transmembrane proteins that are responsible for the fusion of vesicles to their target membranes, which is crucial for vesicle cargo delivery and managing lipid membrane integrity. Several Rab GTPases are thought to regulate these fusion events at target membranes to provide specificity to vesicle trafficking patterns (Novick et al., 2006). We also know that spatial and temporal regulation helps to specialise the complexes that SNAREs form to differentiate trafficking patterns between cellular membranes (Bassham and Blatt, 2008; Enami et al., 2009). With all that we know about how vesicles move around the cell and get to where they need to go, we are still working towards understanding how the delivery and fusion of membranes during vesicle transport are temporally regulated.
 Tomosyn is a Qa-SNARE binding protein that is highly conserved in both plants and animals (Fujita et al., 1998; Li et al 2019). When it is overexpressed in rat neuroendocrine cells, secretion is inhibited, while a knock-out mutant in C. elegans enhances secretion (Gladycheva et al., 2007; Gracheva et al., 2006). Similar to the findings in animals, in this issue of Plant Physiology, Li et al. (2019) provide interesting evidence for how the overexpression of the plant homolog TOMOSYN (AtTMS) in Arabidopsis impairs secretion and plant development by reducing cell plate formation and pollen grain maturation.
Tomosyn is a Qa-SNARE binding protein that is highly conserved in both plants and animals (Fujita et al., 1998; Li et al 2019). When it is overexpressed in rat neuroendocrine cells, secretion is inhibited, while a knock-out mutant in C. elegans enhances secretion (Gladycheva et al., 2007; Gracheva et al., 2006). Similar to the findings in animals, in this issue of Plant Physiology, Li et al. (2019) provide interesting evidence for how the overexpression of the plant homolog TOMOSYN (AtTMS) in Arabidopsis impairs secretion and plant development by reducing cell plate formation and pollen grain maturation.
Looking at the structure of the AtTMS protein goes a long way in explaining its function. Li et al. (2019) report that the protein structure of AtTMS includes conserved N-terminal WD40 repeats and a C-terminal R-SNARE motif, which both point to its function as a protein that can interact with or be a part of the SNARE fusion complex. Proteins with WD40 repeats are classically involved in multi-protein complex assemblies because the rigid WD40 repeats can act as a scaffold that supports protein interactions (Smith et al., 1999). Likewise, the C-terminal R-SNARE motif of AtTMS and its localization to the PM and TGN place it in proximity to where SNARE complexes form.
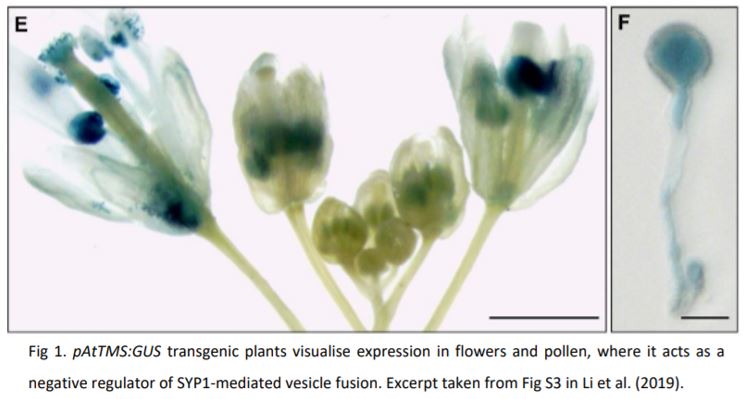
While SNAREs are notoriously promiscuous, they form specialised complexes through spatial and temporal separation. The authors show that AtTMS is expressed in several plant tissues, but in pollen it is mainly expressed at the end of the microspore stage (Fig. 1) and overexpression affects the normal development of the nucleus and cell wall deposition. This developmentally coordinated expression may suggest that AtTMS acts as a brake to regulate the time and location of exocytosis during pollen development. Moreover, yeast-two-hybrid and luciferase complementation assays showed that AtTMS specifically interacted with SNAREs localized to the plasma membrane and trans-Golgi network (TGN), further indicating its function in secretory vesicle trafficking pathways. Although AtTMS is expressed in many tissues, these interactions appear to occur preferentially with SNAREs expressed in pollen, which may indicate a tissue or organ specificity of AtTMS-dependent SNARE regulation. While the authors report that overexpression did affect seedling growth and root hair expansion, this finding may point to ectopic expression effects more than endogenous function. The AtTMS knockout and overexpressing lines produced by the authors provide excellent molecular and genetic tools that will help to further define the activity AtTMS has in different cell types, particularly to resolve the whole-plant phenotypes with the presented protein-interaction data.
Li et al. provide some elegant experiments to show that not only does AtTMS outcompete R-SNAREs for Qa-SNARE binding in vitro, but that overexpression of R-SNAREs can rescue the secretory block imposed by AtTMS overexpression. This is similar to the SNARE-interactor hand-shake that has been described in other SNARE protein complex formations (Karnik et al., 2013), where the interacting proteins of the SNARE fusion complex do so in a sequential pattern during SNARE-mediated secretion events.
References
Bassham, D.C., and Blatt, M.R. (2008). SNAREs: cogs and coordinators in signaling and development. Plant Physiol. 147, 1504–1515.
Cai, H., Reinisch, K., and Ferro-Novick, S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671–682.
Enami, K., Ichikawa, M., Uemura, T., Kutsuna, N., Hasezawa, S., Nakagawa, T., Nakano, A., and Sato, M.H. (2009). Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 50, 280–289.
Fujita, Y., Shirataki, H., Sakisaka, T., Asakura, T., Ohya, T., Kotani, H., Yokoyama, S., Nishioka, H., Matsuura, Y., Mizoguchi, A., et al. (1998). Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20, 905–915.
Gladycheva, S.E., Lam, A.D., Liu, J., D’Andrea-Merrins, M., Yizhar, O., Lentz, S.I., Ashery, U., Ernst, S.A., and Stuenkel, E.L. (2007). Receptor-mediated regulation of tomosyn-syntaxin 1A interactions in bovine adrenal chromaffin cells. J. Biol. Chem. 282, 22887–22899.
Gracheva, E.O., Burdina, A.O., Holgado, A.M., Berthelot-Grosjean, M., Ackley, B.D., Hadwiger, G., Nonet, M.L., Weimer, R.M., and Richmond, J.E. (2006). Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol. 4, e261.
Karnik, R., Grefen, C., Bayne, R., Honsbein, A., Köhler, T., Kioumourtzoglou, D., Williams, M., Bryant, N.J., and Blatt, M.R. (2013). Arabidopsis Sec1/Munc18 Protein SEC11 Is a Competitive and Dynamic Modulator of SNARE Binding and SYP121-Dependent Vesicle Traffic. Plant Cell 25, 1368–1382.
Li, B., Li, Y., Liu, F., Tan, X., Rui, Q., Tong, Y., Qiao, L., Gao, R., Shi, R., Li, Y., Bao, Y. (2019) Overexpression of AtTMS blocks secretion during pollen development by competitively binding to SYP1s. Plant Physiol. https://doi.org/10.1104/pp.19.00965
Novick, P., Medkova, M., Dong, G., Hutagalung, A., Reinisch, K., and Grosshans, B. (2006). Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem. Soc. Trans. 34, 683–686.
Sanderfoot, A.A., Assaad, F., and Raikhel, N. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569.
Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185.




