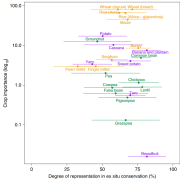
Plant Science Research Weekly: Sept. 22, 2023
Review: Rapid auxin signaling: Unknowns old and new
 You might think you’ve read enough about auxin, but I recommend you take this opportunity to read one more article, this very interesting and enjoyable review by Fielder and Friml. Auxin has figured prominently in both the classical and molecular eras of plant biology. However, the exciting findings of auxin’s transcriptional responses through the TIR1 auxin receptor relatively early in the molecular era really dominated the field over the past 25 years, overshadowing the classically-described “fast” auxin responses that aren’t explained by transcription. Further complications arose when it was revealed that the effects ascribed to Auxin Binding Protein 1 through analysis of the Arabidopsis abp1 mutant were not quite as they seemed. It’s not an overstatement to say, as the authors do, that this finding “poured fuel on the fire of a community that was already, as human nature sometimes dictates, divided by its favoritism for either ABP1 or TIR1 as the more relevant auxin receptor.” That’s enough foreshadowing, so please turn to the article for the rest of this elegantly constructed update. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102443
You might think you’ve read enough about auxin, but I recommend you take this opportunity to read one more article, this very interesting and enjoyable review by Fielder and Friml. Auxin has figured prominently in both the classical and molecular eras of plant biology. However, the exciting findings of auxin’s transcriptional responses through the TIR1 auxin receptor relatively early in the molecular era really dominated the field over the past 25 years, overshadowing the classically-described “fast” auxin responses that aren’t explained by transcription. Further complications arose when it was revealed that the effects ascribed to Auxin Binding Protein 1 through analysis of the Arabidopsis abp1 mutant were not quite as they seemed. It’s not an overstatement to say, as the authors do, that this finding “poured fuel on the fire of a community that was already, as human nature sometimes dictates, divided by its favoritism for either ABP1 or TIR1 as the more relevant auxin receptor.” That’s enough foreshadowing, so please turn to the article for the rest of this elegantly constructed update. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102443
Review: Genome evolution in plants and the origins of innovation
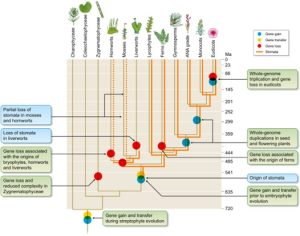 Land plants have enormous diversity; however we do not fully understand how this has arisen. In this review article James Clark discusses how genome dynamics and gene loss contribute to genome evolution and the generation of diversity and complexity. He explains how genome evolution is non-homogenous across time with specific periods where plants underwent rapid evolution. One period is the emergence of land plants, where there was an expansion of pre-existing gene families and the generation of novel gene families to allow the colonization of the land. In this process horizontal gene transfer played an important role, particularly for the evolution of genes involved in stress response. In addition to gene gain, gene loss has had a major influence on plant evolution, particularly within bryophytes. One example is the genes involved in stomata formation, which were present in a bryophyte ancestor but have since been lost in liverworts and some mosses and hornworts. This loss has led to generation of phenotypic diversity for instance the development of the novel gas exchange structure, the air pore, in liverworts. Hence, Clark argues that gene loss plays a significant role in generating diversity and needs to be carefully considered in plant evolution. (Summary by Rose McNully @Rose_McN) New Phytol. 10.1111/nph.19242
Land plants have enormous diversity; however we do not fully understand how this has arisen. In this review article James Clark discusses how genome dynamics and gene loss contribute to genome evolution and the generation of diversity and complexity. He explains how genome evolution is non-homogenous across time with specific periods where plants underwent rapid evolution. One period is the emergence of land plants, where there was an expansion of pre-existing gene families and the generation of novel gene families to allow the colonization of the land. In this process horizontal gene transfer played an important role, particularly for the evolution of genes involved in stress response. In addition to gene gain, gene loss has had a major influence on plant evolution, particularly within bryophytes. One example is the genes involved in stomata formation, which were present in a bryophyte ancestor but have since been lost in liverworts and some mosses and hornworts. This loss has led to generation of phenotypic diversity for instance the development of the novel gas exchange structure, the air pore, in liverworts. Hence, Clark argues that gene loss plays a significant role in generating diversity and needs to be carefully considered in plant evolution. (Summary by Rose McNully @Rose_McN) New Phytol. 10.1111/nph.19242
Review: Improving RNA-based crop protection through nanotechnology and insights from cross-kingdom RNA trafficking
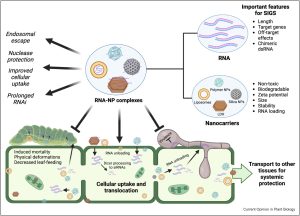 The German physician Paul Ehrlich (not to be confused with the American scientist of the same name) coined the term “magic bullet” (zauberkugel) to describe something that is perfectly and accurately effective. As much as we dream of magic bullets, they are rarely found, but the idea of using spray-on RNA as an agent of pest control seems pretty close. Here, Chen et al. review progress in this field, specifically spray-induced gene silencing (SIGS), and particularly the use of nanotechnology to improve its efficacy. The potential for small RNAs to specifically and effectively silence target genes has been recognized for decades; the challenges in pest control are largely about delivery and stability. The authors review examples of naturally occurring cross-kingdom RNAi, particularly the involvement of extracellular vesicles in enabling movement. Delivery and stability are greatly increased by the encapsulation of the RNA into nanoparticles of various design and composition (including layered double hydroxide clay nanosheets, chitosan, carbon-based materials, liposomes, and star polycations). There’s an interesting discussion about how the nanoparticles enter the plant, and translocation within the plant tissues, and efforts to improve these. Before widely adopted, the fate of the nanocarriers in the environment and impacts on non-target organisms needs to be addressed. Nevertheless, the appeal of spraying on droplets of encapsulated, highly targeted small RNAs as a means of crop protection is real. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102441
The German physician Paul Ehrlich (not to be confused with the American scientist of the same name) coined the term “magic bullet” (zauberkugel) to describe something that is perfectly and accurately effective. As much as we dream of magic bullets, they are rarely found, but the idea of using spray-on RNA as an agent of pest control seems pretty close. Here, Chen et al. review progress in this field, specifically spray-induced gene silencing (SIGS), and particularly the use of nanotechnology to improve its efficacy. The potential for small RNAs to specifically and effectively silence target genes has been recognized for decades; the challenges in pest control are largely about delivery and stability. The authors review examples of naturally occurring cross-kingdom RNAi, particularly the involvement of extracellular vesicles in enabling movement. Delivery and stability are greatly increased by the encapsulation of the RNA into nanoparticles of various design and composition (including layered double hydroxide clay nanosheets, chitosan, carbon-based materials, liposomes, and star polycations). There’s an interesting discussion about how the nanoparticles enter the plant, and translocation within the plant tissues, and efforts to improve these. Before widely adopted, the fate of the nanocarriers in the environment and impacts on non-target organisms needs to be addressed. Nevertheless, the appeal of spraying on droplets of encapsulated, highly targeted small RNAs as a means of crop protection is real. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102441
Commentary: Time to fight the over-hype
 A year ago, graduate student Merritt Khaipho-Burch Tweeted a reaction to an article about a gene described as enhancing yield, which led to lively on-line and in-the-lunchroom discussions about how to realistically measure yield, and, maybe more importantly, where to draw the line between potential and promise. Now, Khaipho-Burch and colleagues have published a set of recommendations for researchers, reviewers, and editors, aiming to cast a more realistic and critical view towards research efforts to improve crop yields. Their five recommendations: Standard definition of yield; Broad replication; Farm-like conditions; Appropriate controls; and Priority to genes breeders might have missed. On this last point, the authors state, “If plant breeders have already worked with a gene for decades, it is extremely unlikely that it will suddenly deliver major yield gains.” Needless to say, this is a much-needed commentary that should be discussed at your next lab meeting. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/d41586-023-02895-w
A year ago, graduate student Merritt Khaipho-Burch Tweeted a reaction to an article about a gene described as enhancing yield, which led to lively on-line and in-the-lunchroom discussions about how to realistically measure yield, and, maybe more importantly, where to draw the line between potential and promise. Now, Khaipho-Burch and colleagues have published a set of recommendations for researchers, reviewers, and editors, aiming to cast a more realistic and critical view towards research efforts to improve crop yields. Their five recommendations: Standard definition of yield; Broad replication; Farm-like conditions; Appropriate controls; and Priority to genes breeders might have missed. On this last point, the authors state, “If plant breeders have already worked with a gene for decades, it is extremely unlikely that it will suddenly deliver major yield gains.” Needless to say, this is a much-needed commentary that should be discussed at your next lab meeting. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/d41586-023-02895-w
Microtubule-associated proteins regulate fruit shape in tomato
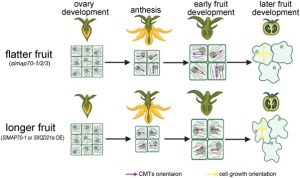 Microtubule binding proteins are important in determining fruit shape by driving changes in microtubule arrangement. However, in depth molecular studies on these in tomato (Solanum lycopersicum) have been limited. Here, Bao et al. investigated the microtubule binding protein Microtubule-Associated Protein 70 (MAP70), which is highly expressed in developing tomato fruits. Knocking out map70-1 and its homolog map70-2 with CRISPR/Cas9 led to flatter tomato fruits due to a reduction in fruit length and width. They labelled microtubules in the map70-1/map70-2 mutant with the microtubule marker GFP-MAP65-1 and used laser scanning confocal microscopy to look at microtubule organisation in endocarp cells. At an individual cell level, the mutant had more-ordered microtubules than the wild type, whereas at the supracellular level microtubule orientation was more varied, resulting in a lack of coordination in the expansion of different cells and hence flatter fruits. Beyond this they used yeast-2-hybrid, and bimolecular complementation to show that MAP70-1 interacts with the scaffold protein IQD21a. This interaction stabilizes microtubules and affects the dynamic rearrangement of microtubules during fruit development. Interestingly, an unrelated paper in Nature Plants by Li et al. found that elongated tomato fruits are more crush-resistant and amenable to mechanical harvesting, with the elongated shape conferred by alterations in cell division patterns. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad231
Microtubule binding proteins are important in determining fruit shape by driving changes in microtubule arrangement. However, in depth molecular studies on these in tomato (Solanum lycopersicum) have been limited. Here, Bao et al. investigated the microtubule binding protein Microtubule-Associated Protein 70 (MAP70), which is highly expressed in developing tomato fruits. Knocking out map70-1 and its homolog map70-2 with CRISPR/Cas9 led to flatter tomato fruits due to a reduction in fruit length and width. They labelled microtubules in the map70-1/map70-2 mutant with the microtubule marker GFP-MAP65-1 and used laser scanning confocal microscopy to look at microtubule organisation in endocarp cells. At an individual cell level, the mutant had more-ordered microtubules than the wild type, whereas at the supracellular level microtubule orientation was more varied, resulting in a lack of coordination in the expansion of different cells and hence flatter fruits. Beyond this they used yeast-2-hybrid, and bimolecular complementation to show that MAP70-1 interacts with the scaffold protein IQD21a. This interaction stabilizes microtubules and affects the dynamic rearrangement of microtubules during fruit development. Interestingly, an unrelated paper in Nature Plants by Li et al. found that elongated tomato fruits are more crush-resistant and amenable to mechanical harvesting, with the elongated shape conferred by alterations in cell division patterns. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad231
Oryza glumaepatula: A wild relative to improve drought tolerance in cultivated rice
 When we speak about rice, we’re often referring to one of two domesticated species, Oryza japonica or Oryza indica. However, there are an additional 25 species in the genus Oryza. These so-called wild relatives harbor substantial genetic diversity that holds promise for crop improvement. Here, Prakash et al. surveyed 22 Oryza species for drought tolerance traits. This study is impressively robust. The authors looked at phenotypes of plants grown in greenhouse cylinders and screenhouse paddies in both well-watered and drought-stress conditions, and measured a wide range of growth and physiological outcomes. Their initial screens pointed to O. glumaepatula as showing improved drought tolerance, so they next screened a panel of 69 accessions (obtained from the International Rice Research Institute, IRRI, and the National Institute of Genetics, Japan). The O. glumaepatula panel was subject to genome sequencing analysis and genome-wide association studies to identify regions associated with “least reduction in shoot biomass” and “lower stomatal density” under drought. Interestingly, the drought-tolerant accessions originate from flood-prone regions in the Amazon River Basin, supporting a previous connection between flooding tolerance and drought tolerance, possibly arising from heightened ethylene sensitivity. This impressive effort opens many interesting lines of inquiry. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiad485
When we speak about rice, we’re often referring to one of two domesticated species, Oryza japonica or Oryza indica. However, there are an additional 25 species in the genus Oryza. These so-called wild relatives harbor substantial genetic diversity that holds promise for crop improvement. Here, Prakash et al. surveyed 22 Oryza species for drought tolerance traits. This study is impressively robust. The authors looked at phenotypes of plants grown in greenhouse cylinders and screenhouse paddies in both well-watered and drought-stress conditions, and measured a wide range of growth and physiological outcomes. Their initial screens pointed to O. glumaepatula as showing improved drought tolerance, so they next screened a panel of 69 accessions (obtained from the International Rice Research Institute, IRRI, and the National Institute of Genetics, Japan). The O. glumaepatula panel was subject to genome sequencing analysis and genome-wide association studies to identify regions associated with “least reduction in shoot biomass” and “lower stomatal density” under drought. Interestingly, the drought-tolerant accessions originate from flood-prone regions in the Amazon River Basin, supporting a previous connection between flooding tolerance and drought tolerance, possibly arising from heightened ethylene sensitivity. This impressive effort opens many interesting lines of inquiry. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiad485
Bacterial pathogens deliver water- and solute-permeable channels to plant cells
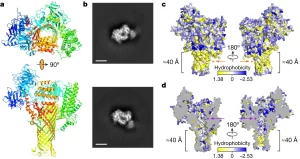 What do you do when you’ve identified a gene that you know is important, but you don’t know how it functions? Usually, you can get hints from homology searches, overexpression studies, or the identification of protein domains, but sometimes those approaches don’t work. That’s where the story of the pathogen virulence protein AvrE from Psueodomonas syringae (and related proteins) sat for a long time. Now, Nomura et al. have cracked the case. They started with AlphaFold2 analysis, the AI system that predicts protein structures. Interestingly, the structural model predicted a “mushroom shaped” protein with a clear central channel. The authors then used several methods to test this model, including cryoEM imaging. Expression in oocytes revealed that the protein creates a functional channel for water and small molecules. Further studies suggested that the channels could be blocked by polyamidoamine dendrimers, and, interestingly, they authors found that these molecules also inhibit the virulence of the pathogens. This exciting paper not only solves a decades-old riddle, but also beautifully illustrates how the combination of AlphaFold2 and cryoEM (both recent advancements) can provide clues to protein function. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-023-06531-5
What do you do when you’ve identified a gene that you know is important, but you don’t know how it functions? Usually, you can get hints from homology searches, overexpression studies, or the identification of protein domains, but sometimes those approaches don’t work. That’s where the story of the pathogen virulence protein AvrE from Psueodomonas syringae (and related proteins) sat for a long time. Now, Nomura et al. have cracked the case. They started with AlphaFold2 analysis, the AI system that predicts protein structures. Interestingly, the structural model predicted a “mushroom shaped” protein with a clear central channel. The authors then used several methods to test this model, including cryoEM imaging. Expression in oocytes revealed that the protein creates a functional channel for water and small molecules. Further studies suggested that the channels could be blocked by polyamidoamine dendrimers, and, interestingly, they authors found that these molecules also inhibit the virulence of the pathogens. This exciting paper not only solves a decades-old riddle, but also beautifully illustrates how the combination of AlphaFold2 and cryoEM (both recent advancements) can provide clues to protein function. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-023-06531-5
From friend to threat: the mutualist-pathogen transition of plant-fungal interaction is determined by a small gene cluster
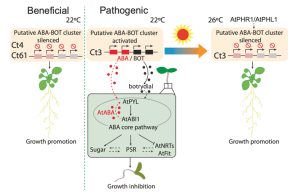 The infection modes of microbes in hosts can be reversible and can range from pathogenic to mutualistic, depending on the environmental conditions. The plant-associated fungus Colletotrichum tofieldiae (Ct) has been shown to promote primary root growth and increase shoot fresh weight of Arabidopsis in phosphate deficit conditions. However, Hiruma et al. identified a strain, Ct3, that acts as a plant pathogen. RNA-seq analysis in Arabidopsis revealed that during the Ct3 pathogenesis, plant ABA-responsive genes and phosphate starvation response (PSR) genes are up-regulated. This suggests that Ct3 represses plant growth by activating the ABA signaling pathway. Further RNA-seq analysis in C. tofieldiae identified a cluster of genes responsible for biosynthesis of ABA and the phytotoxic metabolite botrydial (BOT) in Ct3, which are co-activated during root colonization. Disruption of ABA/BOT biosynthesis genes in Ct3, for example at high temperature, causes the transition of infection mode of Ct3 from pathogenic to beneficial and allows plant growth under low Pi conditions. These results are summarized in their model; unlike the beneficial strains Ct4/Ct61, the ABA/BOT genes in Ct3 are activated during root colonization to produce ABA or related metabolites, which activates the ABA signaling pathway in the host to repress the nutrient acquisition and inhibit plant growth. (Summary by Diwen Wang @Diwen_w) Nature Comms. 10.1038/s41467-023-40867-w
The infection modes of microbes in hosts can be reversible and can range from pathogenic to mutualistic, depending on the environmental conditions. The plant-associated fungus Colletotrichum tofieldiae (Ct) has been shown to promote primary root growth and increase shoot fresh weight of Arabidopsis in phosphate deficit conditions. However, Hiruma et al. identified a strain, Ct3, that acts as a plant pathogen. RNA-seq analysis in Arabidopsis revealed that during the Ct3 pathogenesis, plant ABA-responsive genes and phosphate starvation response (PSR) genes are up-regulated. This suggests that Ct3 represses plant growth by activating the ABA signaling pathway. Further RNA-seq analysis in C. tofieldiae identified a cluster of genes responsible for biosynthesis of ABA and the phytotoxic metabolite botrydial (BOT) in Ct3, which are co-activated during root colonization. Disruption of ABA/BOT biosynthesis genes in Ct3, for example at high temperature, causes the transition of infection mode of Ct3 from pathogenic to beneficial and allows plant growth under low Pi conditions. These results are summarized in their model; unlike the beneficial strains Ct4/Ct61, the ABA/BOT genes in Ct3 are activated during root colonization to produce ABA or related metabolites, which activates the ABA signaling pathway in the host to repress the nutrient acquisition and inhibit plant growth. (Summary by Diwen Wang @Diwen_w) Nature Comms. 10.1038/s41467-023-40867-w
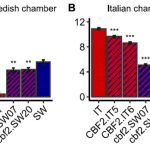
A large effect genetic trade-off is caused by a single mutation in CBF2
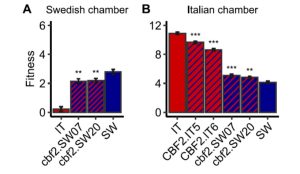 Understanding the genetic basis of local adaptation of a species is an important but thorny problem. Now that whole genomes are readily characterized, it’s not hard to see lots of differences between populations, but pulling meaning and demonstrating functional consequences out of those differences is very difficult. Here, Lee et al. convincingly show that the well-known cold-responsive transcription factor CBF2 has both positive and negative fitness effects, in a context-dependent manner. Previous studies have shown that initiating the cold acclimation response when it isn’t beneficial is detrimental to plant growth. In this work, the authors compared the impact of a naturally-occurring CBF2 polymorphism on fitness in Sweden (where cold acclimation is necessary) and Italy (where it is not), using near-isogenic lines (NILs) outdoors and gene-editing plants in growth chambers. The authors note that the NIL lines include larger genomic regions and several polymorphisms besides CBF2, but the NIL results are corroborated by the gene-edited plants (which can’t be grown outdoors in the EU). The authors recorded the fitness of hundreds of individual plants, using per-plant fruit number as a measure of fitness as it encompasses both survival and fecundity. Their results showed that at both locations, plants with the “wrong” CBF2 allele fared significantly worse than those with the right allele. Interestingly but not surprisingly, the “wrong allele” plants in Sweden took the biggest hit on survival (death by cold when acclimation doesn’t occur), whereas the “wrong allele” plants in Italy took the biggest hit on fecundity (with the precise mechanism to be determined). (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.09.11.557195
Understanding the genetic basis of local adaptation of a species is an important but thorny problem. Now that whole genomes are readily characterized, it’s not hard to see lots of differences between populations, but pulling meaning and demonstrating functional consequences out of those differences is very difficult. Here, Lee et al. convincingly show that the well-known cold-responsive transcription factor CBF2 has both positive and negative fitness effects, in a context-dependent manner. Previous studies have shown that initiating the cold acclimation response when it isn’t beneficial is detrimental to plant growth. In this work, the authors compared the impact of a naturally-occurring CBF2 polymorphism on fitness in Sweden (where cold acclimation is necessary) and Italy (where it is not), using near-isogenic lines (NILs) outdoors and gene-editing plants in growth chambers. The authors note that the NIL lines include larger genomic regions and several polymorphisms besides CBF2, but the NIL results are corroborated by the gene-edited plants (which can’t be grown outdoors in the EU). The authors recorded the fitness of hundreds of individual plants, using per-plant fruit number as a measure of fitness as it encompasses both survival and fecundity. Their results showed that at both locations, plants with the “wrong” CBF2 allele fared significantly worse than those with the right allele. Interestingly but not surprisingly, the “wrong allele” plants in Sweden took the biggest hit on survival (death by cold when acclimation doesn’t occur), whereas the “wrong allele” plants in Italy took the biggest hit on fecundity (with the precise mechanism to be determined). (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.09.11.557195
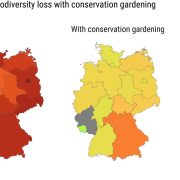
Putting conservation gardening on the map
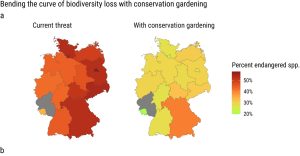 We’re living in a time of increasing awareness of environmental degradation, but this awareness is often not accompanied by opportunities for direct actions. Here, Munschek et al. describe conservation gardening (growing endangered native plants) as a strategy by which people can support local biodiversity. As the authors point out, native plants often require less water and fertilizer than conventional garden plants, can provide better opportunities for pollinators, and have “substantial potential to catalyze transformative change in bending the curve of biodiversity loss”. To move people from wanting to do something to doing something, information and access are needed, which is why this is such an innovative project. Focusing on Germany as a case study, the authors looked at all plants on the red list per federal state, identified those that could be amenable to cultivation in private or community spaces (gardens, parks, even balconies), and listed them alongside their site requirements and how to find seeds or seedlings in an easy-to-use database (https://conservation-gardening.shinyapps.io/app-en/). What a brilliant plan. And as the authors say, the more demand there is for gardening with native, endangered species, the more likely that suppliers will supply these plants. Providing people with a simple and direct action isn’t going to solve the biodiversity crisis, but it can have an impact, and, more importantly, provide some hope to these sometimes hopeless-feeling times. (Summary by Mary Williams @PlantTeaching) Sci. Rep. 10.1038/s41598-023-39432-8
We’re living in a time of increasing awareness of environmental degradation, but this awareness is often not accompanied by opportunities for direct actions. Here, Munschek et al. describe conservation gardening (growing endangered native plants) as a strategy by which people can support local biodiversity. As the authors point out, native plants often require less water and fertilizer than conventional garden plants, can provide better opportunities for pollinators, and have “substantial potential to catalyze transformative change in bending the curve of biodiversity loss”. To move people from wanting to do something to doing something, information and access are needed, which is why this is such an innovative project. Focusing on Germany as a case study, the authors looked at all plants on the red list per federal state, identified those that could be amenable to cultivation in private or community spaces (gardens, parks, even balconies), and listed them alongside their site requirements and how to find seeds or seedlings in an easy-to-use database (https://conservation-gardening.shinyapps.io/app-en/). What a brilliant plan. And as the authors say, the more demand there is for gardening with native, endangered species, the more likely that suppliers will supply these plants. Providing people with a simple and direct action isn’t going to solve the biodiversity crisis, but it can have an impact, and, more importantly, provide some hope to these sometimes hopeless-feeling times. (Summary by Mary Williams @PlantTeaching) Sci. Rep. 10.1038/s41598-023-39432-8