Plant Science Research Weekly: October 9, 2020
Review: Less is more, natural loss-of-function mutation is a strategy for adaptation
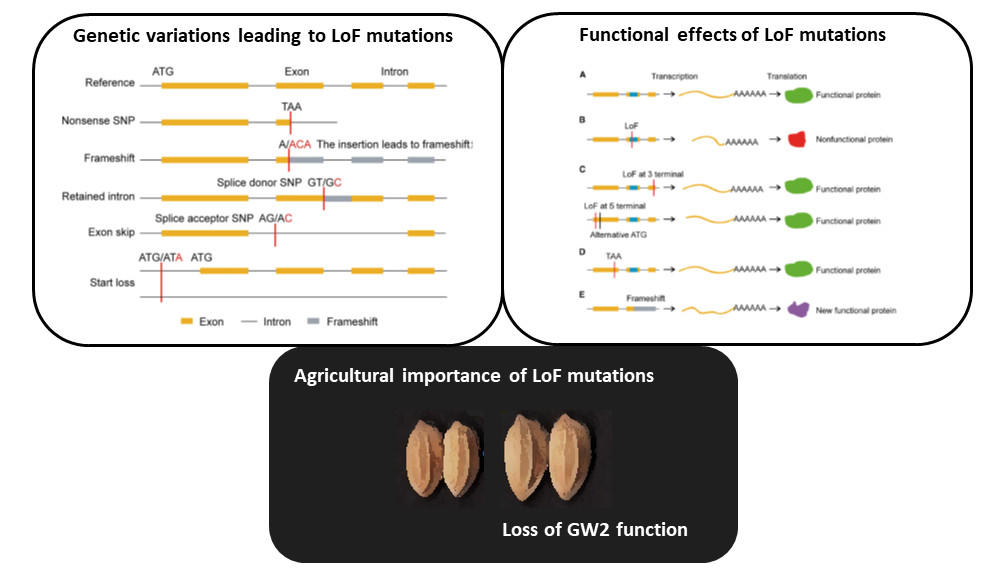
Gene gain through duplication and gene loss through loss-of-function (LoF) mutations determine genetic variation underlying diversification and adaptive evolution. In this review, Xu and Guo highlight the importance of LoF mutations in the evolutionary success of several organisms as revealed by advancements in whole-genome sequencing. The authors focused on the genome-wide distribution of LoF variants in natural populations. Many deleterious LoF mutations show low allele frequencies and tend to accumulate in redundant genes belonging to large families (as paralogs can take over their function). Similar genome-wide analyses also provide information on the “essential gene set” of an organism: absence of deleterious LoF variants at a given gene suggests its crucial role for survival. Nevertheless, not all the LoF mutations lead to complete knockouts because the functional effect also depends on their location. Moreover, LoF mutations could generate truncated proteins with a dominant-negative effect or new functional proteins with similar features. In essence, LoF variants seem to have a great impact on adaptation of the species to new environments. Supporting evidence comes from genetic studies in Petunia: LoF mutations in genes controlling flower scent and color determined changes in pollinator attraction. The authors suggest boosting this approach in crops: novel LoF mutations could be employed as valuable source of genetic variation in breeding programs. For example, LoF variants in Grain Width 2 (GW2) have already been used to increase grain yield in rice. To conclude, gene loss could also be beneficial as proposed by the LESS IS MORE hypothesis. (Summary and image adaptation by Michela Osnato @michela_osnato) Plant Communications 10.1016/j.xplc.2020.100103
Review: Applications of CRISPR–Cas in agriculture and plant biotechnology
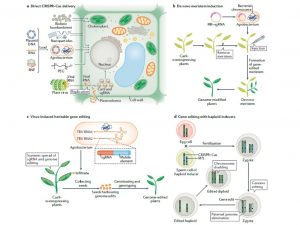 This week’s announcement of the Nobel Prize in Chemistry being awarded to Emmanuelle Charpentier and Jennifer A. Doudna “for the development of a method for genome editing” comes as no surprise to many. CRISPR (clustered regularly interspaced short palindromic repeats)– Cas (CRISPR-associated protein) is a phage immunity system in archaea and bacteria. The system is used to induce a DNA double-strand break (DSB) at a target site and this technology has revolutionized genetic manipulation of crops by targeted and precise editing of the genome. This review by Zhu et al. discusses the state of the art of this technology. CRISPR–Cas technology can give rise to new genotypes by deletion of negative elements that are responsible for traits that are harmful to the plant and it can be used for introducing mutation at targeted sites that can help the plant gain functions. Some of the examples discussed are yield increase in rice by editing C terminus of Oryza sativa LOGL5, which encodes a cytokinin-activation enzyme, increasing quality of wheat by targeting conserved regions of gluten genes and developing gluten free wheat, induction of disease resistance in rice by mutating the promoter region of O. sativa SWEET11 which can give resistance to X. oryzae pv. Oryzae. Plant breeding application in generating male sterility and haploid induction is also discussed, as is its use in accelerating the domestication of crops like quinoa and wheatgrass. Future prospects will include new methods of delivery and organelle genome editing. (Summary by Arun K. Shanker @arunshanker) Nature Rev. Mol. Cell Biol. 10.1038/s41580-020-00288-9
This week’s announcement of the Nobel Prize in Chemistry being awarded to Emmanuelle Charpentier and Jennifer A. Doudna “for the development of a method for genome editing” comes as no surprise to many. CRISPR (clustered regularly interspaced short palindromic repeats)– Cas (CRISPR-associated protein) is a phage immunity system in archaea and bacteria. The system is used to induce a DNA double-strand break (DSB) at a target site and this technology has revolutionized genetic manipulation of crops by targeted and precise editing of the genome. This review by Zhu et al. discusses the state of the art of this technology. CRISPR–Cas technology can give rise to new genotypes by deletion of negative elements that are responsible for traits that are harmful to the plant and it can be used for introducing mutation at targeted sites that can help the plant gain functions. Some of the examples discussed are yield increase in rice by editing C terminus of Oryza sativa LOGL5, which encodes a cytokinin-activation enzyme, increasing quality of wheat by targeting conserved regions of gluten genes and developing gluten free wheat, induction of disease resistance in rice by mutating the promoter region of O. sativa SWEET11 which can give resistance to X. oryzae pv. Oryzae. Plant breeding application in generating male sterility and haploid induction is also discussed, as is its use in accelerating the domestication of crops like quinoa and wheatgrass. Future prospects will include new methods of delivery and organelle genome editing. (Summary by Arun K. Shanker @arunshanker) Nature Rev. Mol. Cell Biol. 10.1038/s41580-020-00288-9
SNAP ‘n’ Track: Protein localization using fluorescent dyes
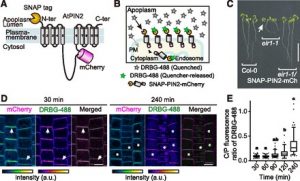 Proteins’ sub-cellular localizations provide a wealth of information regarding their functional attributes. Protein localization in plant cells is usually done through genetically combining fluorescent proteins to the protein-of-interest. Now, Iwatate and colleagues report the successful localization of target proteins in planta by adding to the protein of interest a tag that is subsequently covalently attached to a synthetic dye, in an approach termed O6-alkylguanine-DNA alkyltransferase (SNAP) tagging. The authors employed different commercial SNAP dyes to label tagged microtubules in stably transformed tobacco BY-2 cells and Arabidopsis seedlings and found them to efficiently track microtubule changes during cell division. To examine polar protein localization, the authors tracked the endocytosis of auxin efflux transporter PIN2 using DRBG-488 dye and mCherry tag and found that the dye faithfully labels PIN2 and could track its internalization. They also found that, as shown previously, freshly synthesized PIN2 proteins are preferentially deposited at the cell plate during cell division in Arabidopsis roots. This report, thus, confirms the veracity of a powerful protein tagging technique in plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Cell 10.1105/tpc.20.00439
Proteins’ sub-cellular localizations provide a wealth of information regarding their functional attributes. Protein localization in plant cells is usually done through genetically combining fluorescent proteins to the protein-of-interest. Now, Iwatate and colleagues report the successful localization of target proteins in planta by adding to the protein of interest a tag that is subsequently covalently attached to a synthetic dye, in an approach termed O6-alkylguanine-DNA alkyltransferase (SNAP) tagging. The authors employed different commercial SNAP dyes to label tagged microtubules in stably transformed tobacco BY-2 cells and Arabidopsis seedlings and found them to efficiently track microtubule changes during cell division. To examine polar protein localization, the authors tracked the endocytosis of auxin efflux transporter PIN2 using DRBG-488 dye and mCherry tag and found that the dye faithfully labels PIN2 and could track its internalization. They also found that, as shown previously, freshly synthesized PIN2 proteins are preferentially deposited at the cell plate during cell division in Arabidopsis roots. This report, thus, confirms the veracity of a powerful protein tagging technique in plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Cell 10.1105/tpc.20.00439
PlaCCI (Plant cell cycle indicator); fluorescent sensor for spatiotemporal cell cycle analysis
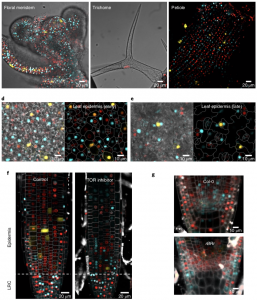 The cell cycle requires a series of transitions from G1 to S, S to G2, G2 to M and M to G1 phases. Determining the cell cycle phase is critical for understanding the molecular events specific to a given stage of cell cycle. However, markers for identifying these cell cycle transitions in plants are not well established. Here, Desvoyes et al. designed and evaluated cell-cycle specific markers in Arabidopsis. The authors used CDT1a (replication initiator protein)-CFP, H3.1 (Histone protein)-mRFP, and CYCB1;1-GFP as markers for G1, S + early G2, and late G2 + M phase respectively. The markers were individually evaluated in independent transgenic lines; however, these individual reporters are not amenable for analyzing all phases of cell cycle as one has to combine these lines genetically. To make it handier, the authors designed all three reporters in the same construct and named it plant cell cycle indicator (PlaCCI). PlaCCI allows live imaging of cell cycle markers in various organs like roots, leaf, petiole, meristem and trichomes. The authors observed that PlaCCI doesn’t affect the cellular growth, meristem size, or enoreduplication status and can be used with various drug treatments and mutant background. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/ s41477-020-00770-4
The cell cycle requires a series of transitions from G1 to S, S to G2, G2 to M and M to G1 phases. Determining the cell cycle phase is critical for understanding the molecular events specific to a given stage of cell cycle. However, markers for identifying these cell cycle transitions in plants are not well established. Here, Desvoyes et al. designed and evaluated cell-cycle specific markers in Arabidopsis. The authors used CDT1a (replication initiator protein)-CFP, H3.1 (Histone protein)-mRFP, and CYCB1;1-GFP as markers for G1, S + early G2, and late G2 + M phase respectively. The markers were individually evaluated in independent transgenic lines; however, these individual reporters are not amenable for analyzing all phases of cell cycle as one has to combine these lines genetically. To make it handier, the authors designed all three reporters in the same construct and named it plant cell cycle indicator (PlaCCI). PlaCCI allows live imaging of cell cycle markers in various organs like roots, leaf, petiole, meristem and trichomes. The authors observed that PlaCCI doesn’t affect the cellular growth, meristem size, or enoreduplication status and can be used with various drug treatments and mutant background. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/ s41477-020-00770-4
Haplotype-resolved genome analyses of a heterozygous diploid potato
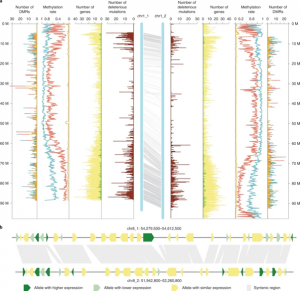 Potato (Solanum tuberosum) is the third most important food crop worldwide. It is also a clonally propagated tetraploid, making progress in breeding genetically improved cultivars extremely difficult. Therefore, there are ongoing efforts to redomesticate potato from a tetraploid, tuber-propagated crop into one that is diploid, inbred-line based, and propagated by seed. However, to do so an improved understanding of the genome landscape of potato is essential. Here, Zhou et al. used various cutting-edge sequencing technologies and a selfing population to generate a haplotype-resolved assembly of a diploid potato. Haplotypes are the distinct sequences of the haploid chromosomes. Most genome assemblies consist of an artificial consensus of both haplotypes, whereas a haplotype-resolved genome considers the different alleles that exist for the same genes, allowing for allele-specific analysis. The authors used two approaches to de novo assemble the potato genome, and eventually combined the results to generate a comprehensive genome of 24 pseudochromosomes. Deleterious mutations and tightly linked genes were identified that will be important targets for future gene editing or molecular selection approaches in this important species. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0699-x
Potato (Solanum tuberosum) is the third most important food crop worldwide. It is also a clonally propagated tetraploid, making progress in breeding genetically improved cultivars extremely difficult. Therefore, there are ongoing efforts to redomesticate potato from a tetraploid, tuber-propagated crop into one that is diploid, inbred-line based, and propagated by seed. However, to do so an improved understanding of the genome landscape of potato is essential. Here, Zhou et al. used various cutting-edge sequencing technologies and a selfing population to generate a haplotype-resolved assembly of a diploid potato. Haplotypes are the distinct sequences of the haploid chromosomes. Most genome assemblies consist of an artificial consensus of both haplotypes, whereas a haplotype-resolved genome considers the different alleles that exist for the same genes, allowing for allele-specific analysis. The authors used two approaches to de novo assemble the potato genome, and eventually combined the results to generate a comprehensive genome of 24 pseudochromosomes. Deleterious mutations and tightly linked genes were identified that will be important targets for future gene editing or molecular selection approaches in this important species. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0699-x
The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth
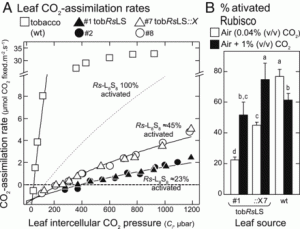 The rate of carboxylation by Rubisco versus the rate of the competing oxygenation reaction limits photosynthesis in some conditions, so many researchers are investigating ways to enhance Rubisco. Rubisco is not limited to green plants but can be found in other lineages including red algae and photosynthetic bacteria other than cyanobacteria. The “red” Rubisco (found in red algae and some proteobacteria) are characterized by a higher affinity for the carboxylation reaction, and so provide an attractive route for photosynthetic enhancement. Here, Gunn et al. introduced into tobacco and E. coli a codon-optimized red Rubisco operon from the photosynthetic bacterium Rhodobacter sphaeroides and showed that the enzyme (a type IC Rubisco) is correctly assembled and functional. The addition of the bacteria’s Rubisco activase enzyme (a metabolic repair chaperon) increased Rubisco activity even more. The fact that similar results were found in E. coli and tobacco indicates that products of directed evolution can be prescreened in the simpler expression system. These findings provide new avenues or the development of Rubisco enhancement in crop plants. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2011641117
The rate of carboxylation by Rubisco versus the rate of the competing oxygenation reaction limits photosynthesis in some conditions, so many researchers are investigating ways to enhance Rubisco. Rubisco is not limited to green plants but can be found in other lineages including red algae and photosynthetic bacteria other than cyanobacteria. The “red” Rubisco (found in red algae and some proteobacteria) are characterized by a higher affinity for the carboxylation reaction, and so provide an attractive route for photosynthetic enhancement. Here, Gunn et al. introduced into tobacco and E. coli a codon-optimized red Rubisco operon from the photosynthetic bacterium Rhodobacter sphaeroides and showed that the enzyme (a type IC Rubisco) is correctly assembled and functional. The addition of the bacteria’s Rubisco activase enzyme (a metabolic repair chaperon) increased Rubisco activity even more. The fact that similar results were found in E. coli and tobacco indicates that products of directed evolution can be prescreened in the simpler expression system. These findings provide new avenues or the development of Rubisco enhancement in crop plants. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2011641117
Transcriptional regulation of PLETHORA1 in the root meristem through an importin and its two antagonistic cargos

Plant root growth is sustained by stem cells in the root meristems, and the stem cell fate is maintained by the quiescent center (QC). PLETHORA (PLT) protein gradients contribute to maintaining the root meristem cell fate, although the transcriptional regulatory mechanism that establishes the PLT gradient is still in question. To address this, Xiong et al. studied JANUS, one of the transcriptional regulators of PLT1. By downregulating JANUS transcription, the authors demonstrated its role on root meristem activity through transcriptional regulation of PLTs. Additionally, the author studied a JANUS-interacting Pol II, NRPB10, and its occupancy at the promoter region of PLT1. This suggested that JANUS activates the transcription of PLTs by recruiting RNA pol II to the PLT1 promoter. The authors also showed the involvement of the IMB4 importin (a protein that facilitates the movement of other proteins into the nucleus) in nuclear accumulation of JANUS. Finally, the authors showed the competitive interaction of GIF1 a (negative transcriptional regulator of PLTs) with JANUS. The authors suggest two antagonistic PLT regulators, GIF1 and JANUS, compete for same importin for nuclear accumulation and that this contributes to the spatiotemporal regulation of the transcription of PLT1 to its gradient during root development. (Summary by Sunita Pathak @psunita980) Plant Cell 10.1105/tpc.20.00108
Combinatorial engineering of signaling networks for drought tolerance
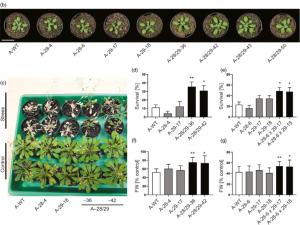 Several potential targets have been suggested to improve drought tolerance and water-use efficiency of crops, but many genes when upregulated can cause negative growth trade-offs. Stress recognition and signaling proteins are attractive targets as they may exert control over multiple downstream pathways and are only activated during stress so are less likely to cause negative phenotypes. Schulz et al. performed novel combinatorial engineering with calcium-dependent protein kinases (CPKs) to identify gene combination that increased drought tolerance. They used 15 Arabidopsis CPK genes to transform Nicotiana tabacum with random combinations of CPKs and screened the progeny for drought resistance and negative growth phenotypes. CPK28 and CPK29 were identified through the screen to increase drought tolerance despite neither being previously implicated in abiotic stress. Further analysis in both Arabidopsis and tobacco uncovered that transformation with both CPK28 and CPK29 are jointly required for increased drought tolerance, possibly through enhanced brassinosteroid signaling. The combinatorial approach used in this study can be used to uncover hidden synergistic interactions between genes in complex genetic networks for both basic and applied research. (Summary by Katy Dunning @plantmomkaty) Plant Biotechnol. J. 10.1111/pbi.13441
Several potential targets have been suggested to improve drought tolerance and water-use efficiency of crops, but many genes when upregulated can cause negative growth trade-offs. Stress recognition and signaling proteins are attractive targets as they may exert control over multiple downstream pathways and are only activated during stress so are less likely to cause negative phenotypes. Schulz et al. performed novel combinatorial engineering with calcium-dependent protein kinases (CPKs) to identify gene combination that increased drought tolerance. They used 15 Arabidopsis CPK genes to transform Nicotiana tabacum with random combinations of CPKs and screened the progeny for drought resistance and negative growth phenotypes. CPK28 and CPK29 were identified through the screen to increase drought tolerance despite neither being previously implicated in abiotic stress. Further analysis in both Arabidopsis and tobacco uncovered that transformation with both CPK28 and CPK29 are jointly required for increased drought tolerance, possibly through enhanced brassinosteroid signaling. The combinatorial approach used in this study can be used to uncover hidden synergistic interactions between genes in complex genetic networks for both basic and applied research. (Summary by Katy Dunning @plantmomkaty) Plant Biotechnol. J. 10.1111/pbi.13441