Plant Science Research Weekly: October 8, 2021
Review: 3D electron microscopy for imaging organelles in plants and algae
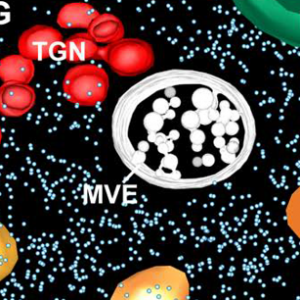 Have you noticed that the quality and resolution of cell images has been getting better and better? Weiner et al. review the recent advances in 3D electron microscopy (EM) technologies that have provided these strikingly beautiful and informative images. Like “classic” transmission EM, EM tomography requires the production of thin sections, either through cryo-preservation (Cryo-EM) or embedding in plastic resin. The sections are imaged from multiple angles by tilting the sample under the electron beam, and then 3D reconstructions are calculated. An alternative approach is serial block-face scanning EM (SBF-SEM). Here, an embedded sample is imaged by scanning EM, then an extremely thin layer removed by an ultramicrotome, and the sample is imaged again. Alternatively, the thin layer is removed by milling with a focused ion beam (FIB-SEM). In correlative light and EM (CLEM), EM methods are combined with light-based imaging, for example fluorescence. This review describes sample preparation required for each of these methods and provides examples of how they have been used in plant cell biology. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab449
Have you noticed that the quality and resolution of cell images has been getting better and better? Weiner et al. review the recent advances in 3D electron microscopy (EM) technologies that have provided these strikingly beautiful and informative images. Like “classic” transmission EM, EM tomography requires the production of thin sections, either through cryo-preservation (Cryo-EM) or embedding in plastic resin. The sections are imaged from multiple angles by tilting the sample under the electron beam, and then 3D reconstructions are calculated. An alternative approach is serial block-face scanning EM (SBF-SEM). Here, an embedded sample is imaged by scanning EM, then an extremely thin layer removed by an ultramicrotome, and the sample is imaged again. Alternatively, the thin layer is removed by milling with a focused ion beam (FIB-SEM). In correlative light and EM (CLEM), EM methods are combined with light-based imaging, for example fluorescence. This review describes sample preparation required for each of these methods and provides examples of how they have been used in plant cell biology. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab449
Review: Illuminating the hidden world of calcium ions in plants with a universe of indicators
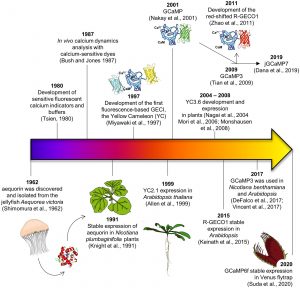 Calcium ions (Ca2+) are ubiquitous and essential signals, with a very steep concentration gradient from outside to inside the cell. Numerous studies from across the kingdoms have demonstrated that transient Ca2+ fluxes convey information of many types. Here, Grenzi et al. provide an Update on the tools that have been developed to quantify and localize these all-important calcium signals, including an interesting timeline of their development and applications. Calcium sensors fall into two camps: dyes and genetically encoded calcium indicators (GECIs). A drawback of the dyes is their need to be introduced into cells, but they also do not require the generation of transgenic plants. Much of this update addresses the pros and cons of various GECIs, including the cameleon indicators that use FRET (Förster resonance energy transfer) to measure the calcium-mediated interaction between a calcium binding protein calmodulin (CaM) and a peptide derived from one of its interacting partners. When calcium is present, the proximity of this protein/peptide pair reorients to closer proximity, allowing fluorescence transfer. These indicators are ratiometric, as they can be normalized to between emission wavelenths to determine expression-level calcium levels. If you’re at all interested in knowing more about how this very important messenger is quantified in plants, have a look. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab339
Calcium ions (Ca2+) are ubiquitous and essential signals, with a very steep concentration gradient from outside to inside the cell. Numerous studies from across the kingdoms have demonstrated that transient Ca2+ fluxes convey information of many types. Here, Grenzi et al. provide an Update on the tools that have been developed to quantify and localize these all-important calcium signals, including an interesting timeline of their development and applications. Calcium sensors fall into two camps: dyes and genetically encoded calcium indicators (GECIs). A drawback of the dyes is their need to be introduced into cells, but they also do not require the generation of transgenic plants. Much of this update addresses the pros and cons of various GECIs, including the cameleon indicators that use FRET (Förster resonance energy transfer) to measure the calcium-mediated interaction between a calcium binding protein calmodulin (CaM) and a peptide derived from one of its interacting partners. When calcium is present, the proximity of this protein/peptide pair reorients to closer proximity, allowing fluorescence transfer. These indicators are ratiometric, as they can be normalized to between emission wavelenths to determine expression-level calcium levels. If you’re at all interested in knowing more about how this very important messenger is quantified in plants, have a look. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab339
Review: The long road to engineering durable disease resistance in wheat
 This review by Wulff and Krattinger is a beautifully written “must read”. As the title suggests, it takes the reader on a journey of scientific progress, starting from the “first controlled cross between two wheat species” to the present, with a look into the future. Triticum aestivum (bread wheat) is a critically important food crop with an absurdly complex genome (5x that of the human genome) and, thanks to its long history of domestication, ample genetic diversity. Diseases and pests destroy enough grain to bake “290 billion loaves of bread” per year, or if my math is correct, more than half a loaf of bread per person per week. The review covers the discovery, diversity, and function of resistance (R) genes, as well as more recent efforts to describe the complete R-gene atlas. The rapidly evolving technological innovations that have enabled these efforts are described in a way that is understandable even to a non-specialist. Finally, the steps that must be taken to obtain durable disease resistance, through stacking of multiple R genes alongside broad-spectrum resistance genes, are outlined. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Biotechnol. 10.1016/j.copbio.2021.09.002
This review by Wulff and Krattinger is a beautifully written “must read”. As the title suggests, it takes the reader on a journey of scientific progress, starting from the “first controlled cross between two wheat species” to the present, with a look into the future. Triticum aestivum (bread wheat) is a critically important food crop with an absurdly complex genome (5x that of the human genome) and, thanks to its long history of domestication, ample genetic diversity. Diseases and pests destroy enough grain to bake “290 billion loaves of bread” per year, or if my math is correct, more than half a loaf of bread per person per week. The review covers the discovery, diversity, and function of resistance (R) genes, as well as more recent efforts to describe the complete R-gene atlas. The rapidly evolving technological innovations that have enabled these efforts are described in a way that is understandable even to a non-specialist. Finally, the steps that must be taken to obtain durable disease resistance, through stacking of multiple R genes alongside broad-spectrum resistance genes, are outlined. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Biotechnol. 10.1016/j.copbio.2021.09.002
Review: Cycling in a crowd: coordination of plant cell division, growth and cell fate
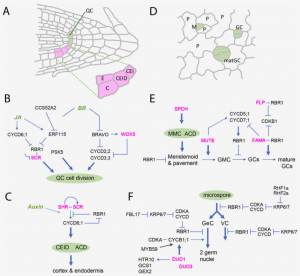 Unlike animal cells, plant cells cannot migrate due to the presence of interconnected rigid cell walls, thus plant development relies on a continuous supply of new cells, (fueled by the cell cycle) to form new organs throughout their life. This excellent review by Sablowski and Gutierrez focuses on the intricate regulatory networks and cell cycle key players that integrate cell growth and shape, metabolic activity, cell division and morphogenesis, while at the same time highlighting key questions regarding the missing links between mechanical stress, cell division orientation and chromatin accessibility during cell cycle progression, among other interesting promising research avenues. Furthermore, the authors spotlight the Retinoblastoma pathway, a regulatory module that continues to appear as a central node coordinating different proliferation and differentiation processes mainly through its repressor activity and highly specific physical interactions with master developmental regulators. The authors conclude that although much progress has been made in the plant development field, most of this progress is based on Arabidopsis research, therefore, in order understand the vast diversity of shapes that characterizes plants it will be essential to investigate developmental tool kits through an evo-devo (evolutionary developmental biology) lens. (Summary by Jesus Leon @jesussaur) Plant Cell 10.1093/plcell/koab222
Unlike animal cells, plant cells cannot migrate due to the presence of interconnected rigid cell walls, thus plant development relies on a continuous supply of new cells, (fueled by the cell cycle) to form new organs throughout their life. This excellent review by Sablowski and Gutierrez focuses on the intricate regulatory networks and cell cycle key players that integrate cell growth and shape, metabolic activity, cell division and morphogenesis, while at the same time highlighting key questions regarding the missing links between mechanical stress, cell division orientation and chromatin accessibility during cell cycle progression, among other interesting promising research avenues. Furthermore, the authors spotlight the Retinoblastoma pathway, a regulatory module that continues to appear as a central node coordinating different proliferation and differentiation processes mainly through its repressor activity and highly specific physical interactions with master developmental regulators. The authors conclude that although much progress has been made in the plant development field, most of this progress is based on Arabidopsis research, therefore, in order understand the vast diversity of shapes that characterizes plants it will be essential to investigate developmental tool kits through an evo-devo (evolutionary developmental biology) lens. (Summary by Jesus Leon @jesussaur) Plant Cell 10.1093/plcell/koab222
Seed biologists beware: End-of-trial estimation of initial viability may be error-prone
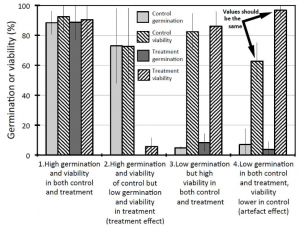 Assessing the viability of ungerminated seeds at the end of a trial is a common practice in seed germination experiments. By doing this, researchers aim to establish the number of viable seeds that were sown in an experiment and estimate germination parameters accordingly. Still, can we be sure that those seeds found to be unviable at the end of the experiment were always unviable? Here, Lamont and colleagues show us that we have reasons to be skeptical. For end-of-trial estimations of seed viability to be reliable, they should match those carried at the beginning. However, 16 out of 40 Leucadendron species exposed to different treatments exhibited different viability values depending on the assessment time, even in controls where viability changes were unexpected. These differences could not be explained by aging, given that cold-stored seeds exhibited minimal viability reduction. Therefore, seeds were assumed to have died during the experiment. Interestingly, seeds with low germination rates seemed to be more susceptible to this artifact, suggesting that the longer a seed remains ungerminated, the higher the probability it dies during the trial. Given this, this research provides fundamental guidelines about the use and proper interpretation of viability assessments in seed germination biology. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Preprints 10.20944/preprints202109.0256.v1
Assessing the viability of ungerminated seeds at the end of a trial is a common practice in seed germination experiments. By doing this, researchers aim to establish the number of viable seeds that were sown in an experiment and estimate germination parameters accordingly. Still, can we be sure that those seeds found to be unviable at the end of the experiment were always unviable? Here, Lamont and colleagues show us that we have reasons to be skeptical. For end-of-trial estimations of seed viability to be reliable, they should match those carried at the beginning. However, 16 out of 40 Leucadendron species exposed to different treatments exhibited different viability values depending on the assessment time, even in controls where viability changes were unexpected. These differences could not be explained by aging, given that cold-stored seeds exhibited minimal viability reduction. Therefore, seeds were assumed to have died during the experiment. Interestingly, seeds with low germination rates seemed to be more susceptible to this artifact, suggesting that the longer a seed remains ungerminated, the higher the probability it dies during the trial. Given this, this research provides fundamental guidelines about the use and proper interpretation of viability assessments in seed germination biology. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Preprints 10.20944/preprints202109.0256.v1
Ten simple rules for choosing a PhD supervisor
 Choosing a PhD supervisor is one of the most important and difficult decisions of an academic career. This new entry by Jabre et al. in the “Ten simple rules” series endeavors to highlight some of the factors to consider when faced with this decision. These span from the early steps of identifying potential supervisors based on their research interests and methods, to how to investigate if the adivisor, research group, and institution are good fits for your interest. Talking to people who know you and can advise is crucial, as is talking to the individual’s current and former students. Lab cultures vary tremendously, and no matter how much you love the research question, you’ll be unhappy if your approach to work and decision making is at odds with the lab group you find yourself in. This is a very useful set of “rules” to share with those who are preparing their search for a PhD supervisor – please share! (Summary by Mary Williams @PlantTeaching) PLOS Comput. Biol. 10.1371/journal.pcbi.1009330
Choosing a PhD supervisor is one of the most important and difficult decisions of an academic career. This new entry by Jabre et al. in the “Ten simple rules” series endeavors to highlight some of the factors to consider when faced with this decision. These span from the early steps of identifying potential supervisors based on their research interests and methods, to how to investigate if the adivisor, research group, and institution are good fits for your interest. Talking to people who know you and can advise is crucial, as is talking to the individual’s current and former students. Lab cultures vary tremendously, and no matter how much you love the research question, you’ll be unhappy if your approach to work and decision making is at odds with the lab group you find yourself in. This is a very useful set of “rules” to share with those who are preparing their search for a PhD supervisor – please share! (Summary by Mary Williams @PlantTeaching) PLOS Comput. Biol. 10.1371/journal.pcbi.1009330
Lessons on textile history and fibre durability from a 4,000-year-old Egyptian flax yarn
 “Food, fuel, and fiber” sometimes is used to describe our utter dependence on plants, but often, other than cotton, the “fiber” part is under appreciated. However, there is a growing interest in bio-based materials, both for their greater recyclability and as an alternative to petroleum-derived materials (see ‘Biobased Composites for Mobility and Transportation: Moving towards Sustainability’). Flax (Linum spp.) has been a source of linen fibers for thousands of years. In a recent article, Melelli et al. used modern microscopy methods to compare the fibers from contemporary linen to those from a 4000 year old Egyptian cloth in order to investigate how the fibers aged and compare their microscopic structures. These methods included scanning electron microscopy (SEM), nano-tomography, second harmonic generation (SHG) imaging (which highlights crystalline cellulose in the cell walls), and atomic force microscopy tests in peak force quantitative nano-mechanical (AFM-PF-QNM). Collectively these approaches showed the skill with which the ancient Egyptians extracted the fibers, and noticeable differences between ancient and modern samples between the fiber diameter and lumen size. Importantly, the studies also demonstrate the extreme durability of the flax plant cell walls, supporting their ability to be used in strong and durable “next-generation environmentally friendly composite materials”. I wonder what tools will be available to examine these new materials 4000 years from now? (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-021-00998-8
“Food, fuel, and fiber” sometimes is used to describe our utter dependence on plants, but often, other than cotton, the “fiber” part is under appreciated. However, there is a growing interest in bio-based materials, both for their greater recyclability and as an alternative to petroleum-derived materials (see ‘Biobased Composites for Mobility and Transportation: Moving towards Sustainability’). Flax (Linum spp.) has been a source of linen fibers for thousands of years. In a recent article, Melelli et al. used modern microscopy methods to compare the fibers from contemporary linen to those from a 4000 year old Egyptian cloth in order to investigate how the fibers aged and compare their microscopic structures. These methods included scanning electron microscopy (SEM), nano-tomography, second harmonic generation (SHG) imaging (which highlights crystalline cellulose in the cell walls), and atomic force microscopy tests in peak force quantitative nano-mechanical (AFM-PF-QNM). Collectively these approaches showed the skill with which the ancient Egyptians extracted the fibers, and noticeable differences between ancient and modern samples between the fiber diameter and lumen size. Importantly, the studies also demonstrate the extreme durability of the flax plant cell walls, supporting their ability to be used in strong and durable “next-generation environmentally friendly composite materials”. I wonder what tools will be available to examine these new materials 4000 years from now? (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-021-00998-8



