Plant Science Research Weekly: October 7th, 2022
Genome-edited rice deficient in two 4-COUMARATE:COENZYME A LIGASE genes displays diverse lignin alterations
 Lignin is one of the most important end-products of the cinnamate/monolignol pathway and it is abundant in the secondary cell wall of vascular plants. In grasses, lignins are derived from monolignols, p-hydroxycinnamates, and a flavonoid tricin. In the proposed cinnamate/monolignol pathway, 4-COUMARATE:COENZYME A LIGASE (4CL) modulates the pathway flux that converts hydroxycinnamic acids (such as p-coumaric acid) into the products. In the rice genome there are five 4CL isoforms, of which Os4CL3 and Os4CL4 have been suggested to be more relevant than other 4CLs in lignin biosynthesis. In this work, Afifi et al. describe how Os4CL3 and Os4CL4 isoforms have overlapping but divergent roles during the production of lignin monomers in rice. They generated different CRIPSR/Cas9 rice mutant lines for Os4CL3 and Os4CL4 and then they analyzed the different cell wall chemical structures. The results suggest that while Os4CL3 might be involved mainly in the production of guaiacyl units and to a lesser extent in the syringyl units, Os4CL4 alone plays a role in tricin biosynthesis. While both mutant lines presented deficiencies in lignin polymer units, each line showed specific characteristics regarding the production of lignin. Concluding, this work proposes new targets for breeding to optimize grass biomass for the sustainable production of biochemicals. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac450
Lignin is one of the most important end-products of the cinnamate/monolignol pathway and it is abundant in the secondary cell wall of vascular plants. In grasses, lignins are derived from monolignols, p-hydroxycinnamates, and a flavonoid tricin. In the proposed cinnamate/monolignol pathway, 4-COUMARATE:COENZYME A LIGASE (4CL) modulates the pathway flux that converts hydroxycinnamic acids (such as p-coumaric acid) into the products. In the rice genome there are five 4CL isoforms, of which Os4CL3 and Os4CL4 have been suggested to be more relevant than other 4CLs in lignin biosynthesis. In this work, Afifi et al. describe how Os4CL3 and Os4CL4 isoforms have overlapping but divergent roles during the production of lignin monomers in rice. They generated different CRIPSR/Cas9 rice mutant lines for Os4CL3 and Os4CL4 and then they analyzed the different cell wall chemical structures. The results suggest that while Os4CL3 might be involved mainly in the production of guaiacyl units and to a lesser extent in the syringyl units, Os4CL4 alone plays a role in tricin biosynthesis. While both mutant lines presented deficiencies in lignin polymer units, each line showed specific characteristics regarding the production of lignin. Concluding, this work proposes new targets for breeding to optimize grass biomass for the sustainable production of biochemicals. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac450
Tobacco leaf tissue rapidly detoxifies direct salt loads without activation of calcium and SOS signaling
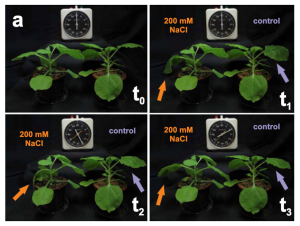 Salinity stress is one of the primary abiotic causes of crop loss worldwide. In roots, the early response to high salt levels is coordinated largely via the well characterized salt-overly sensitive (SOS) pathway, which is dependent on Ca2+ signaling. However, how plants cope with elevated salt levels in leaves is less clear. Here, Graus and Li et al. infiltrated salt into leaves of Nicotiana benthamiana to study this aspect of the salinity response. Treatment with 200mM NaCl resulted in immediate loss of turgor, reflected by a gradual down turning of leaves. However, this turgor loss was short lived, with leaves returning to their original positions within 150 minutes of treatment. This recovery cycle was also observed in photosynthesis performance, with the initial reduction in photosynthetic rates resolving in a similar timespan. Using qPCR, the researchers also showed that there are no transcriptional changes in ABA or SOS signaling pathways, or proline synthesis pathways during this NaCl treatment. By using the Ca2+ sensor GCaMP3 and the Ca2+ channel blocker lanthanum chloride, the group also showed that unlike in roots, the leaf response to salinity stress is not dependent on Ca2+ signaling. Finally, the group overexpressed the cation/H+ transporter AtNHX1, revealing that it is also responsible for the salinity response in leaves, although unlike in roots it is not regulated by cytosolic Ca2+ levels. (Summary by Rory Burke @rorby95) New Phytol. 10.1111/nph.18501
Salinity stress is one of the primary abiotic causes of crop loss worldwide. In roots, the early response to high salt levels is coordinated largely via the well characterized salt-overly sensitive (SOS) pathway, which is dependent on Ca2+ signaling. However, how plants cope with elevated salt levels in leaves is less clear. Here, Graus and Li et al. infiltrated salt into leaves of Nicotiana benthamiana to study this aspect of the salinity response. Treatment with 200mM NaCl resulted in immediate loss of turgor, reflected by a gradual down turning of leaves. However, this turgor loss was short lived, with leaves returning to their original positions within 150 minutes of treatment. This recovery cycle was also observed in photosynthesis performance, with the initial reduction in photosynthetic rates resolving in a similar timespan. Using qPCR, the researchers also showed that there are no transcriptional changes in ABA or SOS signaling pathways, or proline synthesis pathways during this NaCl treatment. By using the Ca2+ sensor GCaMP3 and the Ca2+ channel blocker lanthanum chloride, the group also showed that unlike in roots, the leaf response to salinity stress is not dependent on Ca2+ signaling. Finally, the group overexpressed the cation/H+ transporter AtNHX1, revealing that it is also responsible for the salinity response in leaves, although unlike in roots it is not regulated by cytosolic Ca2+ levels. (Summary by Rory Burke @rorby95) New Phytol. 10.1111/nph.18501
Plant immunity is wired differently in different plants
 In plants, microbial invasion is sensed by membrane-bound and cytoplasmic receptors. Our knowledge on the immune signaling that follows microbial recognition is mostly derived from studies on Arabidopsis thaliana. In this plant, receptor activation triggers immune responses that are distinct for each subgroup of receptors. However, recent findings using this model plant have shown that signaling from membrane-bound pattern recognition receptors (PRRs) and cytoplasmic TIR-domain containing NLR-type receptors (TNLs) is linked through the function of the lipase-like protein EDS1. Studies in the solanaceous plant N. benthamiana suggest that in this plant EDS1 works slightly differently that the one in Arabidopsis. In a recent report, Zönnchen et al. looked at functions of PRRs and TNLs in N. benthamiana. Their results suggest that, as for its Arabidopsis counterpart, EDS1 in N. benthamiana is required for TNL signaling, although an intact catalytic triad is dispensable in both plants. Zönnchen et al also found that in contrast to the Arabidopsis system, EDS1 in N. benthamiana seems not to be involved in immune signalling of PRRs. This comparative study highlights the diversity of plant immune responses. (Summary by Mariana Schuster @MariSchuster) New Phytol 10.1111/nph.18511
In plants, microbial invasion is sensed by membrane-bound and cytoplasmic receptors. Our knowledge on the immune signaling that follows microbial recognition is mostly derived from studies on Arabidopsis thaliana. In this plant, receptor activation triggers immune responses that are distinct for each subgroup of receptors. However, recent findings using this model plant have shown that signaling from membrane-bound pattern recognition receptors (PRRs) and cytoplasmic TIR-domain containing NLR-type receptors (TNLs) is linked through the function of the lipase-like protein EDS1. Studies in the solanaceous plant N. benthamiana suggest that in this plant EDS1 works slightly differently that the one in Arabidopsis. In a recent report, Zönnchen et al. looked at functions of PRRs and TNLs in N. benthamiana. Their results suggest that, as for its Arabidopsis counterpart, EDS1 in N. benthamiana is required for TNL signaling, although an intact catalytic triad is dispensable in both plants. Zönnchen et al also found that in contrast to the Arabidopsis system, EDS1 in N. benthamiana seems not to be involved in immune signalling of PRRs. This comparative study highlights the diversity of plant immune responses. (Summary by Mariana Schuster @MariSchuster) New Phytol 10.1111/nph.18511
Arabidopsis latent virus 1, a comovirus widely spread in Arabidopsis thaliana collections
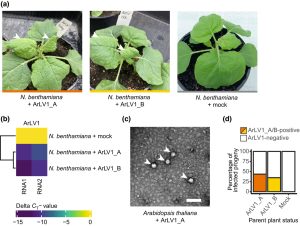 Arabidopsis thaliana has been established as a versatile and important model plant species, with abundant genetic and genomic resources. Through RNA sequencing, Verhoeven et al. identified an unexpected and previously unnoticed latent comovirus (the name “como” derives from cowpea mosaic virus) lurking in the genome of this well-studied plant, which was detected in both laboratory and wild strains. The authors named it Arabidopsis latent virus 1, as it is frequently causes no observable disease symptoms. Although many genomes contain evidence of long-gone viruses in the form of transposons, in this case the virus retains the ability to infect other plants and is seed transmissible to progeny. The authors showed both de novo infection of other Arabidopsis plants and Nicotiana benthamiana, the latter of which showed symptoms of leaf mottling. Although phenotypic consequences of the infection were minimal, mild drought resistance was detected in both species. The authors speculate that this virus is likely to be widespread and have caused “as yet unknown effects on plant performance in a substantial number of studies.” (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.18466
Arabidopsis thaliana has been established as a versatile and important model plant species, with abundant genetic and genomic resources. Through RNA sequencing, Verhoeven et al. identified an unexpected and previously unnoticed latent comovirus (the name “como” derives from cowpea mosaic virus) lurking in the genome of this well-studied plant, which was detected in both laboratory and wild strains. The authors named it Arabidopsis latent virus 1, as it is frequently causes no observable disease symptoms. Although many genomes contain evidence of long-gone viruses in the form of transposons, in this case the virus retains the ability to infect other plants and is seed transmissible to progeny. The authors showed both de novo infection of other Arabidopsis plants and Nicotiana benthamiana, the latter of which showed symptoms of leaf mottling. Although phenotypic consequences of the infection were minimal, mild drought resistance was detected in both species. The authors speculate that this virus is likely to be widespread and have caused “as yet unknown effects on plant performance in a substantial number of studies.” (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.18466
High humidity compromises plant immunity while promoting pathogen growth
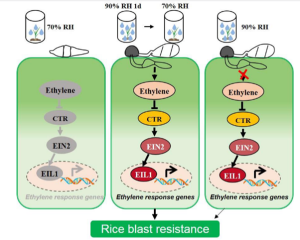 Humidity is considered a key component for the growth of fungal plant pathogens. For many infection assays we either maintain plants in high humidity, or wrap the infected area and keep spraying it with water. This is generally performed to facilitate fungal growth and the assumption that this may not have any effect on plant immunity. However, a recent publication by Qiu et al. indicates that high humidity does affect plant immunity. Qiu et al. performed rice infection assays at moderate (70%) and high (90%) moisture conditions. The authors observed higher humidity leads to faster conidial germination and appressoria formation in Magnaporthe oryzae (rice blast fungus) leading to more severe blast infection in rice. But authors also observed that at higher humidity levels, plants show less severe immune responses when treated with elicitors of immunity or with M. oryzae. Transcriptome analysis indicated reduced expression of ethylene response-related genes at high humidity. Using ethylene signaling mutants and exogenous application of ethylene signaling inhibitors or ethylene analogues, the authors concluded that the reduced defense response at elevated humidity is due to compromised ethylene signaling at high humidity. These findings help us understand why plants are more prone to infections during high rainfall. Additionally, this information may also help farmers strategize disease control strategies based on rainfall. (Summary by Kamal Kumar Malukani @KamalMalukani) Plant Cell Environ. 10.1111/pce.14452
Humidity is considered a key component for the growth of fungal plant pathogens. For many infection assays we either maintain plants in high humidity, or wrap the infected area and keep spraying it with water. This is generally performed to facilitate fungal growth and the assumption that this may not have any effect on plant immunity. However, a recent publication by Qiu et al. indicates that high humidity does affect plant immunity. Qiu et al. performed rice infection assays at moderate (70%) and high (90%) moisture conditions. The authors observed higher humidity leads to faster conidial germination and appressoria formation in Magnaporthe oryzae (rice blast fungus) leading to more severe blast infection in rice. But authors also observed that at higher humidity levels, plants show less severe immune responses when treated with elicitors of immunity or with M. oryzae. Transcriptome analysis indicated reduced expression of ethylene response-related genes at high humidity. Using ethylene signaling mutants and exogenous application of ethylene signaling inhibitors or ethylene analogues, the authors concluded that the reduced defense response at elevated humidity is due to compromised ethylene signaling at high humidity. These findings help us understand why plants are more prone to infections during high rainfall. Additionally, this information may also help farmers strategize disease control strategies based on rainfall. (Summary by Kamal Kumar Malukani @KamalMalukani) Plant Cell Environ. 10.1111/pce.14452



