Plant Science Research Weekly: October 27, 2023
Review: Development of organs for nutrient uptake in parasitic plants and root nodule symbiosis
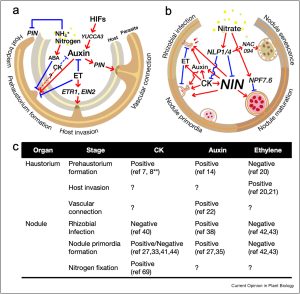 This review by Cui et al. makes the interesting comparison between the developmental processes involved in root nodule formation and haustoria formation by roots of parasitic plants. As the authors observe, both are organs that are produced for the purpose of nutrient acquisition through “intimate relationships” with other organisms, and initiation of each starts with the perception of signals released by the partner (haustorium-inducing factors or Nod factors). In the article, the authors elaborate further on the comparison between these two pathways of organogenesis. For example, both types of organ are derived from lateral roots, and formation of both is suppressed by environmental nitrogen and enhanced by the action of auxin and cytokinins. Finally, the authors suggest that a better understanding of these processes could provide opportunities to engineer synthetic organs. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102473
This review by Cui et al. makes the interesting comparison between the developmental processes involved in root nodule formation and haustoria formation by roots of parasitic plants. As the authors observe, both are organs that are produced for the purpose of nutrient acquisition through “intimate relationships” with other organisms, and initiation of each starts with the perception of signals released by the partner (haustorium-inducing factors or Nod factors). In the article, the authors elaborate further on the comparison between these two pathways of organogenesis. For example, both types of organ are derived from lateral roots, and formation of both is suppressed by environmental nitrogen and enhanced by the action of auxin and cytokinins. Finally, the authors suggest that a better understanding of these processes could provide opportunities to engineer synthetic organs. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102473
Review: Complementing model species with model clades
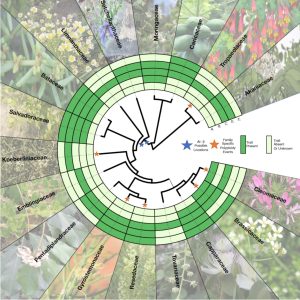 Without doubt, Arabidopsis thaliana has thoroughly demonstrated its usefulness as a model species. In this interesting article by Mabry et al. (with an impressive author list!), the authors propose to expand the Arabidopsis toolkit to encompass its entire order – its family of families, the Brassicales. Brassicales includes approximately 4700 species in 18 families, including the familiar Brassicaceae family (cabbages and Arabidopsis) but also Tropaeolum (nasturtiums), Caricaceae (papaya) and many more. Building a model clade of this magnitude seems ambitious, but current sequencing technologies render it feasible. In addition to the challenge of gathering genomic data that spans the many species and families, there is the additional challenge of extensive polyploidization across the order, which not only increases genome size but also leads to selective gene loss, thus making it more difficult to identify direct comparisons between genes; interestingly, several tools have been developed for addressing these challenges. The article touches on some of the biological insights (flower development, small RNAs) gleaned from Arabidopsis research and how they can be informed by comparative studies on the broader, “model clade”. There’s also discussion about looking at specialized metabolites, “woodiness”, photosynthesis, and extremophytism in the Brasicales. The article concludes with the need for an integrated, global community to work together to expand from model species to model clade. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koad260
Without doubt, Arabidopsis thaliana has thoroughly demonstrated its usefulness as a model species. In this interesting article by Mabry et al. (with an impressive author list!), the authors propose to expand the Arabidopsis toolkit to encompass its entire order – its family of families, the Brassicales. Brassicales includes approximately 4700 species in 18 families, including the familiar Brassicaceae family (cabbages and Arabidopsis) but also Tropaeolum (nasturtiums), Caricaceae (papaya) and many more. Building a model clade of this magnitude seems ambitious, but current sequencing technologies render it feasible. In addition to the challenge of gathering genomic data that spans the many species and families, there is the additional challenge of extensive polyploidization across the order, which not only increases genome size but also leads to selective gene loss, thus making it more difficult to identify direct comparisons between genes; interestingly, several tools have been developed for addressing these challenges. The article touches on some of the biological insights (flower development, small RNAs) gleaned from Arabidopsis research and how they can be informed by comparative studies on the broader, “model clade”. There’s also discussion about looking at specialized metabolites, “woodiness”, photosynthesis, and extremophytism in the Brasicales. The article concludes with the need for an integrated, global community to work together to expand from model species to model clade. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koad260
Review: Challenges to improving plant growth through introduced microbes
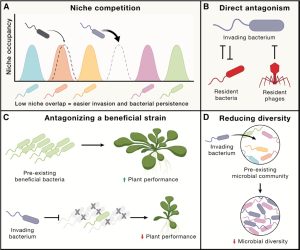 Plants are closely associated with large numbers of microbes that live in, on, and around them; these are collectively called the plant microbiota. Microbes can be pathogenic, neutral, or beneficial. Beneficial microbes might enhance nutrient uptake by the plant or suppress pathogenic microbes. There is growing interest in using introduced microbes to support plant growth, but this effort is complicated by many factors. The plant defense system is of course a big challenge, but going beyond that, interactions between microbial communities provide further difficulties. The new species needs to compete with pre-existing species, and conflicts may arise that harm not only the new ones but also other beneficial microbes. Plants apply selective pressure to the microbial communities, fostering the development of microbiomes through specific immune responses to exclude pathogenesis. The plant immune system regulates microbial balance in various manners, including hormonal signaling, immune receptor signaling and secondary metabolite production. Environmental fluctuations also pose challenges to the deployment of microbial strategies, especially factors like drought, iron deficiencies, and phosphate levels. The crosstalk between the plant immune system and other pathways, such as symbiosis and abiotic stress, further complicates this process. Although questions about how to enhance plant productivity through beneficial microbes remain, our understanding is rapidly expanding, with technologies like high-throughput assays being utilized to identify microbial traits. Innovative strategies, like using engineered microbes to carry several beneficial traits, remain to be tested in real-world scenarios. (Summary by Diwen Wang @Diwen_w) Cell 10.1016/j.cell.2023.08.035
Plants are closely associated with large numbers of microbes that live in, on, and around them; these are collectively called the plant microbiota. Microbes can be pathogenic, neutral, or beneficial. Beneficial microbes might enhance nutrient uptake by the plant or suppress pathogenic microbes. There is growing interest in using introduced microbes to support plant growth, but this effort is complicated by many factors. The plant defense system is of course a big challenge, but going beyond that, interactions between microbial communities provide further difficulties. The new species needs to compete with pre-existing species, and conflicts may arise that harm not only the new ones but also other beneficial microbes. Plants apply selective pressure to the microbial communities, fostering the development of microbiomes through specific immune responses to exclude pathogenesis. The plant immune system regulates microbial balance in various manners, including hormonal signaling, immune receptor signaling and secondary metabolite production. Environmental fluctuations also pose challenges to the deployment of microbial strategies, especially factors like drought, iron deficiencies, and phosphate levels. The crosstalk between the plant immune system and other pathways, such as symbiosis and abiotic stress, further complicates this process. Although questions about how to enhance plant productivity through beneficial microbes remain, our understanding is rapidly expanding, with technologies like high-throughput assays being utilized to identify microbial traits. Innovative strategies, like using engineered microbes to carry several beneficial traits, remain to be tested in real-world scenarios. (Summary by Diwen Wang @Diwen_w) Cell 10.1016/j.cell.2023.08.035
Letter: Finding umbrella trees: Cultivating inclusion and more than survival in a post-affirmative action academy
 In this Letter to the Editor, Dr. Beronda Montgomery discusses umbrella trees (Musanga cecropioides) as metaphors for how to sustain progress after disruption, specifically the decision by the US Supreme Court to ban the use of race-conscious affirmative action in college admissions. Umbrella trees spring up in disrupted spaces and provide support when resources are scarce. As she writes, “To continue progress and mitigate attrition, new ‘umbrella trees’ that address racial representation at the systems level will need to be established and cultivated. These will require us to acknowledge and enable the active attention and interventions needed to promote equity in science, higher education, and society, more broadly.” Clearly it is time to identify more umbrella trees as we navigate difficult times. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koad261
In this Letter to the Editor, Dr. Beronda Montgomery discusses umbrella trees (Musanga cecropioides) as metaphors for how to sustain progress after disruption, specifically the decision by the US Supreme Court to ban the use of race-conscious affirmative action in college admissions. Umbrella trees spring up in disrupted spaces and provide support when resources are scarce. As she writes, “To continue progress and mitigate attrition, new ‘umbrella trees’ that address racial representation at the systems level will need to be established and cultivated. These will require us to acknowledge and enable the active attention and interventions needed to promote equity in science, higher education, and society, more broadly.” Clearly it is time to identify more umbrella trees as we navigate difficult times. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koad261
Identifying candidates from genome wide association studies using gene orthologs
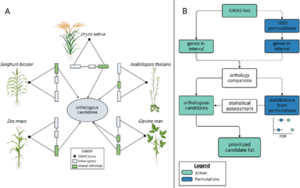 Genome wide association studies (GWAS) identify genomic loci associated with a specific trait. However, these loci often contain many genes, so selecting which to investigate further can be tricky. To improve this Whitt, et al. developed a program called FiReMAGE (filtering results of multi-species, analogous GWAS experiments), which takes loci from many GWAS experiments conducted in different species, identifies genes under each locus, and finds orthologs. Candidate genes are those where the ortholog group is detected in multiple species. Candidate prioritization follows running the pipeline with 1000 random loci and calculating the probability that the locus overlaps with the ortholog by chance. To demonstrate the potential of this program, the authors inputted metal accumulation GWAS data from Arabidopsis thaliana, soybean (Glycine max), rice (Oryza sativa), Sorghum bicolor and maize (Zea mays). By imposing the restriction that the ortholog group had to be identified in all five species, they found 81 strong candidates. Some of these have known roles in metal accumulation, such as the molybdate transport MOT2, whilst others are novel candidates, such as a mevalonate kinase associated with iron homeostasis. Hence, this program provides a powerful way of identifying gene candidates from GWAS loci. (Summary by Rose McNelly @Rose_McN) bioRxiv https://doi.org/10.1101/2023.10.05.561051
Genome wide association studies (GWAS) identify genomic loci associated with a specific trait. However, these loci often contain many genes, so selecting which to investigate further can be tricky. To improve this Whitt, et al. developed a program called FiReMAGE (filtering results of multi-species, analogous GWAS experiments), which takes loci from many GWAS experiments conducted in different species, identifies genes under each locus, and finds orthologs. Candidate genes are those where the ortholog group is detected in multiple species. Candidate prioritization follows running the pipeline with 1000 random loci and calculating the probability that the locus overlaps with the ortholog by chance. To demonstrate the potential of this program, the authors inputted metal accumulation GWAS data from Arabidopsis thaliana, soybean (Glycine max), rice (Oryza sativa), Sorghum bicolor and maize (Zea mays). By imposing the restriction that the ortholog group had to be identified in all five species, they found 81 strong candidates. Some of these have known roles in metal accumulation, such as the molybdate transport MOT2, whilst others are novel candidates, such as a mevalonate kinase associated with iron homeostasis. Hence, this program provides a powerful way of identifying gene candidates from GWAS loci. (Summary by Rose McNelly @Rose_McN) bioRxiv https://doi.org/10.1101/2023.10.05.561051
Using cryo-EM to solve the structures of proteins involved in starch degradation
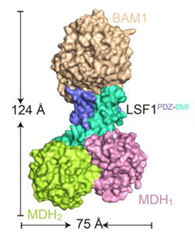 Starch is synthesized in the chloroplasts of leaves during the day and degraded at night. BAM1 (β-AMYLASE 1) catalyzes starch degradation and interacts with the non-catalytic glucan phosphatase called LSF1 (LIKE SEX FOUR 1) and the plastid localised MDH (MALATE DEHYDROGENASE). However, we don’t fully understand how these proteins function together. Here, Liu et al. determined the BAM1-LSF1-MDH structure to 3 Å resolution using cryo-EM. The structure is a dumbbell shape with BAM1 at one end, a MDH dimer at the other end and LSF1 connecting the two. Unfortunately, the DSP (dual specificity domain) and CBM (carbohydrate-binding module) domains of LSF1 were not detected in the structure. Therefore in vitro cross linking coupled with mass spectrometry was used to show that the LSF1 CBM domain was close to the BAM1 catalytic domain. Thus, the LSF1 CBM domain most likely provides polyglucan substrates for BAM1 mediated degradation. The structure also showed that MDH interacts with LSF1 via a latch and gate mechanism that is dependent on three aspartates in MDH. When these Asps are mutated, LSF1 becomes less stable, showing that MDH enhances LSF1 stability. Hence solving the BAM1-LSF1-MDH structure has provided greater insight into the roles of LSF1 and MDH in starch degradation. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad259
Starch is synthesized in the chloroplasts of leaves during the day and degraded at night. BAM1 (β-AMYLASE 1) catalyzes starch degradation and interacts with the non-catalytic glucan phosphatase called LSF1 (LIKE SEX FOUR 1) and the plastid localised MDH (MALATE DEHYDROGENASE). However, we don’t fully understand how these proteins function together. Here, Liu et al. determined the BAM1-LSF1-MDH structure to 3 Å resolution using cryo-EM. The structure is a dumbbell shape with BAM1 at one end, a MDH dimer at the other end and LSF1 connecting the two. Unfortunately, the DSP (dual specificity domain) and CBM (carbohydrate-binding module) domains of LSF1 were not detected in the structure. Therefore in vitro cross linking coupled with mass spectrometry was used to show that the LSF1 CBM domain was close to the BAM1 catalytic domain. Thus, the LSF1 CBM domain most likely provides polyglucan substrates for BAM1 mediated degradation. The structure also showed that MDH interacts with LSF1 via a latch and gate mechanism that is dependent on three aspartates in MDH. When these Asps are mutated, LSF1 becomes less stable, showing that MDH enhances LSF1 stability. Hence solving the BAM1-LSF1-MDH structure has provided greater insight into the roles of LSF1 and MDH in starch degradation. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad259
Listening to the whispers in the air: Plant eavesdropping in action
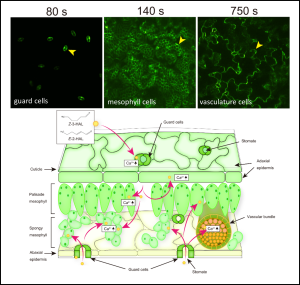 Plants release a variety of volatile organic compounds (VOCs), including green leaf volatiles (GLVs), terpenoids, and amino acid derivatives in response to herbivore damage and injury. Healthy neighboring plants detect these VOCs as warning signals, prompting them to activate defense mechanisms. This process of plant-to-plant communication or plant eavesdropping has been observed several times, but now Aratani et al. were able to define the exact compounds and mechanism via a simple, strategic experimental setup utilizing the transgenic Arabidopsis expressing a calcium biosensor. Real-time changes in the cytosolic calcium levels, [Ca2+]cyt, were observed in the Arabidopsis plants following exposure to VOCs emitted by damaged plants. Among the several VOCs tested in this study, (Z)-3-hexenal (Z-3-HAL) and (E)-2-hexenal (E-2-HAL) caused a rapid and significant increase in [Ca2+]cyt spreading from the tip to the base regions within minute. Exposure to Z-3-HAL and E-2-HAL activated genes related to plant defense mechanisms, such as heat and oxidative stress responses and jasmonic acid signaling. Blocking the increase in [Ca2+]cyt using chemical inhibitors prevented the activation of these defense-related genes, and this effect was reversible. However, when Z-3-HAL was applied directly to a specific plant part, it induced a local increase in [Ca2+]cyt, without systemic propagation to other unstimulated parts. These results suggest that [Ca2+]cyt increases are essential for initiating plant defense responses but they do not trigger long-distance calcium signals within the plant. The rapid changes in [Ca2+]cyt expression in the guard cells, followed by mesophyll and vasculature cells upon Z-3-HAL exposure, was also quantified using tissue-specific high-resolution imaging and mutant analysis, which suggested that VOC (particularly GLV) sensory transduction begins with GLV entry through stomata, subsequently activating plant defense responses in mesophyll and vasculature cells of Arabidopsis leaves. (Summary by Rajarshi Sanyal, @rajarshi_sanyal) Nature Comms. 10.1038/s41467-023-41589-9
Plants release a variety of volatile organic compounds (VOCs), including green leaf volatiles (GLVs), terpenoids, and amino acid derivatives in response to herbivore damage and injury. Healthy neighboring plants detect these VOCs as warning signals, prompting them to activate defense mechanisms. This process of plant-to-plant communication or plant eavesdropping has been observed several times, but now Aratani et al. were able to define the exact compounds and mechanism via a simple, strategic experimental setup utilizing the transgenic Arabidopsis expressing a calcium biosensor. Real-time changes in the cytosolic calcium levels, [Ca2+]cyt, were observed in the Arabidopsis plants following exposure to VOCs emitted by damaged plants. Among the several VOCs tested in this study, (Z)-3-hexenal (Z-3-HAL) and (E)-2-hexenal (E-2-HAL) caused a rapid and significant increase in [Ca2+]cyt spreading from the tip to the base regions within minute. Exposure to Z-3-HAL and E-2-HAL activated genes related to plant defense mechanisms, such as heat and oxidative stress responses and jasmonic acid signaling. Blocking the increase in [Ca2+]cyt using chemical inhibitors prevented the activation of these defense-related genes, and this effect was reversible. However, when Z-3-HAL was applied directly to a specific plant part, it induced a local increase in [Ca2+]cyt, without systemic propagation to other unstimulated parts. These results suggest that [Ca2+]cyt increases are essential for initiating plant defense responses but they do not trigger long-distance calcium signals within the plant. The rapid changes in [Ca2+]cyt expression in the guard cells, followed by mesophyll and vasculature cells upon Z-3-HAL exposure, was also quantified using tissue-specific high-resolution imaging and mutant analysis, which suggested that VOC (particularly GLV) sensory transduction begins with GLV entry through stomata, subsequently activating plant defense responses in mesophyll and vasculature cells of Arabidopsis leaves. (Summary by Rajarshi Sanyal, @rajarshi_sanyal) Nature Comms. 10.1038/s41467-023-41589-9
Mechanism by which viruses are excluded from plant stem cells
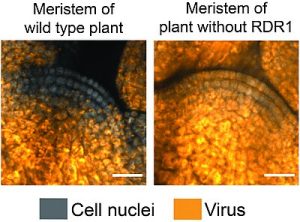 Horticulturalists have long used the technique of meristem culture to propagate plants, as meristems are generally considered to be free of viruses. However, the mechanism by which the stem cells in meristems exclude viruses has been unclear. Here, Incarbone and Bradamante et al. identified roles for RNA-dependent RNA polymerase 1 (RDR1) and salicylic acid (SA) in this mechanism. The authors inoculated leaves with Turnip mosaic virus expressing fluorescent tags, and then used imaging to see if the virus entered the stem cells. Interestingly, in wild-type plants, the virus appears in the stem cells transiently, but is then excluded, whereas in rdr1 mutant plants the virus persists in the stem cells. RDR1 is not itself expressed in stem cells, suggesting it produces a mobile signal that inoculates these cells and protects them from the virus. The authors further identified a role for salicylic acid (SA) in viral exclusion. In viral-treated plants, SA levels increase, and SA promotes the transcription of RDR1. These results suggest a model in which virus infection causes an increase in SA levels, which lead to an increase in RDR1 levels, which amplify antiviral RNAi in infected tissues, providing stem cells with RNA-based virus sequence information that prevents virus proliferation. It remains unclear why this pathway is stem-cell specific. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2302069120
Horticulturalists have long used the technique of meristem culture to propagate plants, as meristems are generally considered to be free of viruses. However, the mechanism by which the stem cells in meristems exclude viruses has been unclear. Here, Incarbone and Bradamante et al. identified roles for RNA-dependent RNA polymerase 1 (RDR1) and salicylic acid (SA) in this mechanism. The authors inoculated leaves with Turnip mosaic virus expressing fluorescent tags, and then used imaging to see if the virus entered the stem cells. Interestingly, in wild-type plants, the virus appears in the stem cells transiently, but is then excluded, whereas in rdr1 mutant plants the virus persists in the stem cells. RDR1 is not itself expressed in stem cells, suggesting it produces a mobile signal that inoculates these cells and protects them from the virus. The authors further identified a role for salicylic acid (SA) in viral exclusion. In viral-treated plants, SA levels increase, and SA promotes the transcription of RDR1. These results suggest a model in which virus infection causes an increase in SA levels, which lead to an increase in RDR1 levels, which amplify antiviral RNAi in infected tissues, providing stem cells with RNA-based virus sequence information that prevents virus proliferation. It remains unclear why this pathway is stem-cell specific. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2302069120

