Plant Science Research Weekly: October 13, 2023
Review. Challenges facing sustainable protein production: Opportunities for cereals
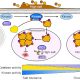
 As the world’s population increases, protein production must increase. If not, levels of protein malnutrition leading to stunted growth, hair loss, and edema will rise. Here Safdar et al. propose that cereals could be bred to be major protein sources. They argue that cereals already make up approximately 50% of our daily calories, so adopting cereals as a protein source would not require a major shift in dietary habits. The largest challenge lies in increasing grain protein content without impacting yield. One promising solution involves the transcription factor HB-2. Increasing HB-2 transcript levels in wheat led to increased hydraulic conductance and greater translocation of nitrogen-rich assimilates to grain. This resulted in approximately 25% more protein in the grains without a loss in yield. The authors also highlight the importance of developing high-throughput, non-destructive methods of assaying protein content, such as hyperspectral imaging, which would allow protein content analysis to be more easily incorporated into breeding programs without requiring excessive time and labor. All in all, increasing protein content in cereals could be a sustainable way of meeting our increasing protein demands. (Summary by Rose McNelly @Rose_McN) Plant Comms. 10.1016/j.xplc.2023.100716
As the world’s population increases, protein production must increase. If not, levels of protein malnutrition leading to stunted growth, hair loss, and edema will rise. Here Safdar et al. propose that cereals could be bred to be major protein sources. They argue that cereals already make up approximately 50% of our daily calories, so adopting cereals as a protein source would not require a major shift in dietary habits. The largest challenge lies in increasing grain protein content without impacting yield. One promising solution involves the transcription factor HB-2. Increasing HB-2 transcript levels in wheat led to increased hydraulic conductance and greater translocation of nitrogen-rich assimilates to grain. This resulted in approximately 25% more protein in the grains without a loss in yield. The authors also highlight the importance of developing high-throughput, non-destructive methods of assaying protein content, such as hyperspectral imaging, which would allow protein content analysis to be more easily incorporated into breeding programs without requiring excessive time and labor. All in all, increasing protein content in cereals could be a sustainable way of meeting our increasing protein demands. (Summary by Rose McNelly @Rose_McN) Plant Comms. 10.1016/j.xplc.2023.100716
Review: Scripting a new dialogue between diazotrophs and crops
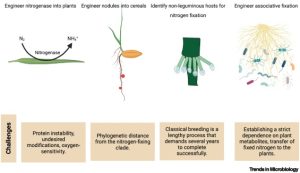 All organisms need nitrogen to produce nucleic and amino acids, but nitrogen-limitation is common for many plants. Although nitrogen is very abundant in the atmosphere, most is inaccessible due to the triple bond that renders N2 relatively inert. Tremendous crop yields in recent decades are attributable largely to the application of ammoinum (NH4+) and other forms of accessible nitrogen to fields, yet these fertilizers are both energetically costly to produce and highly polluting. Diazotrophs are organisms (bacteria or archea) that are able to “fix nitrogen” by breaking the triple bond to produce ammonium, which is accessible for use by other organisms. Some diazotrophs are the familiar endosymbiotic Rhizobia bacteria (which associate with a very limited set of plant species), but others can fix nitrogen as free-living organisms. In this excellent review, Chakraborty et al. explore the challenges and opportunities of exploiting free-living diazotrophs as living fertilizers for crops. A specific focus of this review is the dialogue (information and metabolite exchange) between the plant and the diazotroph. Understanding this dialogue provides opportunities to enhance the production of fixed nitrogen by the diazotroph and its release for uptake by the plant. Finally, the article discusses strategies through which genetically engineered microbes could be employed safely in the field, and in a way that is acceptable to the public. (Summary by Mary Williams @PlantTeaching) Trends Microbiology 10.1016/j.tim.2023.08.007
All organisms need nitrogen to produce nucleic and amino acids, but nitrogen-limitation is common for many plants. Although nitrogen is very abundant in the atmosphere, most is inaccessible due to the triple bond that renders N2 relatively inert. Tremendous crop yields in recent decades are attributable largely to the application of ammoinum (NH4+) and other forms of accessible nitrogen to fields, yet these fertilizers are both energetically costly to produce and highly polluting. Diazotrophs are organisms (bacteria or archea) that are able to “fix nitrogen” by breaking the triple bond to produce ammonium, which is accessible for use by other organisms. Some diazotrophs are the familiar endosymbiotic Rhizobia bacteria (which associate with a very limited set of plant species), but others can fix nitrogen as free-living organisms. In this excellent review, Chakraborty et al. explore the challenges and opportunities of exploiting free-living diazotrophs as living fertilizers for crops. A specific focus of this review is the dialogue (information and metabolite exchange) between the plant and the diazotroph. Understanding this dialogue provides opportunities to enhance the production of fixed nitrogen by the diazotroph and its release for uptake by the plant. Finally, the article discusses strategies through which genetically engineered microbes could be employed safely in the field, and in a way that is acceptable to the public. (Summary by Mary Williams @PlantTeaching) Trends Microbiology 10.1016/j.tim.2023.08.007
Essay: Biomedical publishing: Past, present, and future
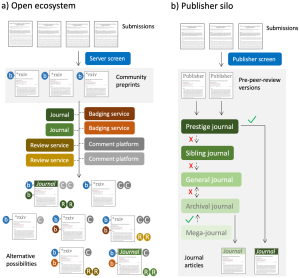 In this essay, Richard Sever (founder of bioRxiv) provides a history of scholarly publishing, starting with the Royal Society Philosophical Transactions (1665). He then provides an overview of the current landscape: not-for-profit society publishers versus for-profit publishers; a breakdown of the real costs incurred by journals; the transition to electronic publishing and archiving; and how calls for open access have disrupted the industry. He next postulates various future directions that publishing and peer review might follow, such as decoupling peer review from dissemination. Although I don’t agree with all his opinions (for example although I support posting preprints, I strongly feel that the process of formal peer review substantially improves most manuscripts), I think it’s important to explore diverse scenarios for the future of academic publishing, bearing in mind the need to be cost effective, equitable, accessible, and reliable. (Summary by Mary Williams @PlantTeaching) PLOS Biology 10.1371/journal.pbio.3002234
In this essay, Richard Sever (founder of bioRxiv) provides a history of scholarly publishing, starting with the Royal Society Philosophical Transactions (1665). He then provides an overview of the current landscape: not-for-profit society publishers versus for-profit publishers; a breakdown of the real costs incurred by journals; the transition to electronic publishing and archiving; and how calls for open access have disrupted the industry. He next postulates various future directions that publishing and peer review might follow, such as decoupling peer review from dissemination. Although I don’t agree with all his opinions (for example although I support posting preprints, I strongly feel that the process of formal peer review substantially improves most manuscripts), I think it’s important to explore diverse scenarios for the future of academic publishing, bearing in mind the need to be cost effective, equitable, accessible, and reliable. (Summary by Mary Williams @PlantTeaching) PLOS Biology 10.1371/journal.pbio.3002234
RNA hairpins underlie preferential use of translation start codons
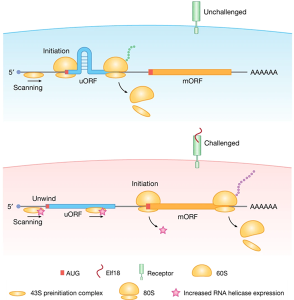 Gene expression regulation facilitates environmental adaptation and survival. This recent paper by Xiang et al. focuses on the regulation of translation, in particular on the mechanism underlying the generation of different translation products in different conditions. In addition to the main start codon (mAUG) and its associated open reading frame (mORF), mRNA molecules can contain upstream start codons (uAUGs) that provide alternative translation start sites that have been shown to be important in stress responses. The authors focused on pathogen-triggered immunity in Arabidopsis thaliana. Many transcripts associated with defense responses contain uAUGs that are bypassed during pathogen response to allow translation from the mAUG. Put in simpler terms, upon immunity response induction, the main AUGs of these environmental stress response genes are chosen for translation, while under normal conditions their uAUGs are preferentially used. The authors found that this change in AUG usage is due to the formation under unstressed conditions of RNA hairpins downstream of uAUGs, which prevent translation from the mAUG. The authors propose that hairpin dissolution is regulated by an increased expression of RNA helicases upon immunity induction, which unwind the RNA hairpin and allow translation of the mORF. Crucially, these helicases have orthologs in yeast and humans, suggesting that this mechanism is common to other eukaryotes. This study tackles a fundamental biological question in Arabidopsis thaliana to reveal a general gene regulation mechanism beyond plants. The authors also provide tools to use this knowledge to design inducible translational reporters to fine-tune gene expression. (Summary by Laura Turchi @turchi_l) Nature 10.1038/s41586-023-06500-y
Gene expression regulation facilitates environmental adaptation and survival. This recent paper by Xiang et al. focuses on the regulation of translation, in particular on the mechanism underlying the generation of different translation products in different conditions. In addition to the main start codon (mAUG) and its associated open reading frame (mORF), mRNA molecules can contain upstream start codons (uAUGs) that provide alternative translation start sites that have been shown to be important in stress responses. The authors focused on pathogen-triggered immunity in Arabidopsis thaliana. Many transcripts associated with defense responses contain uAUGs that are bypassed during pathogen response to allow translation from the mAUG. Put in simpler terms, upon immunity response induction, the main AUGs of these environmental stress response genes are chosen for translation, while under normal conditions their uAUGs are preferentially used. The authors found that this change in AUG usage is due to the formation under unstressed conditions of RNA hairpins downstream of uAUGs, which prevent translation from the mAUG. The authors propose that hairpin dissolution is regulated by an increased expression of RNA helicases upon immunity induction, which unwind the RNA hairpin and allow translation of the mORF. Crucially, these helicases have orthologs in yeast and humans, suggesting that this mechanism is common to other eukaryotes. This study tackles a fundamental biological question in Arabidopsis thaliana to reveal a general gene regulation mechanism beyond plants. The authors also provide tools to use this knowledge to design inducible translational reporters to fine-tune gene expression. (Summary by Laura Turchi @turchi_l) Nature 10.1038/s41586-023-06500-y
Transposable element abundance and fitness in maize
 In this interesting paper, Stritzer et al. asked a simple question, which required a very sophisticated approach to answer. Do transposable elements (TEs) affect fitness in maize? This is a particularly interesting question as 85% of the maize genome is composed of TEs or TE fragments, including over 350,000 TEs. Previous studies have shown a negative impact of TEs on fitness in yeast and Drosophila. A priori, transposons could affect fitness in several ways: through metabolic costs of their replication, through mutational effects caused by transposition, and through impacts on expression levels of nearby genes. The authors examined the US Nested Association Mapping (NAM) population, “a set of 4,975 genotyped recombinant inbred lines (RILs) generated from twenty-five parental inbred lines crossed to the original reference genotype of maize (B73) then self-pollinated for 4-6 generations”. Using grain yield as a proxy for fitness, they found a small negative effect of TE abundance. They further looked at TE position relative to nearby genes, TE age (determined by mutation accumulation from the consensus copy), and TE family. On aggregate, they found that for every additional 14 Mb of TE, fitness was decreased by 1 kernel per plant (about 0.1% of yield). However, younger TEs and TEs closer to genes have a more negative impact. This is an interesting paper with lots more to chew on, have a look. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.09.18.557618
In this interesting paper, Stritzer et al. asked a simple question, which required a very sophisticated approach to answer. Do transposable elements (TEs) affect fitness in maize? This is a particularly interesting question as 85% of the maize genome is composed of TEs or TE fragments, including over 350,000 TEs. Previous studies have shown a negative impact of TEs on fitness in yeast and Drosophila. A priori, transposons could affect fitness in several ways: through metabolic costs of their replication, through mutational effects caused by transposition, and through impacts on expression levels of nearby genes. The authors examined the US Nested Association Mapping (NAM) population, “a set of 4,975 genotyped recombinant inbred lines (RILs) generated from twenty-five parental inbred lines crossed to the original reference genotype of maize (B73) then self-pollinated for 4-6 generations”. Using grain yield as a proxy for fitness, they found a small negative effect of TE abundance. They further looked at TE position relative to nearby genes, TE age (determined by mutation accumulation from the consensus copy), and TE family. On aggregate, they found that for every additional 14 Mb of TE, fitness was decreased by 1 kernel per plant (about 0.1% of yield). However, younger TEs and TEs closer to genes have a more negative impact. This is an interesting paper with lots more to chew on, have a look. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.09.18.557618
A transient approach reduces the time required for transgene-free gene editing
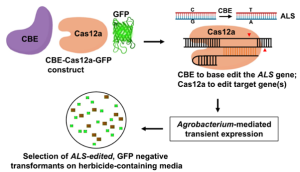 Gene editing in plants is a time-consuming process. One of the challenges of gene editing is to produce transgene-free plants. The transgenes required to carry out selection and editing, such as antibiotic resistance markers and editing enzymes, must be removed, while the edited part of the genome is retained. This selection generally requires 1-3 generations of backcrossing, which takes time, particularly for long-generation species such as perennial plants. Huang et al. have addressed this problem by transiently expressing the gene-editing cassette. Because it is a transient expression, no foreign DNA will get incorporated into the genome. Specifically, the authors transiently expressed a base editor for targeted editing, a target gRNA construct, another gRNA that targets acetolactate synthase, and a GFP construct. While the target site will get a mutation, the other gRNA makes a mutation in acetolactate synthase leading to herbicide tolerance, which serves as a selection marker. Foreign DNA-containing plants show GFP expression, so this provides a negative screenable marker. Using this method, the authors were able to generate transgene-free gene-edited lines in the T0 generation of tomato, tobacco, potato, and citrus. As someone who performed transient assays throughout my PhD, I never imagined such a beautiful application of this technique. (Summary by Kamal Kumar Malukani, @KamalMalukani.) Nature Plants, 10.1038/s41477-023-01520-y.
Gene editing in plants is a time-consuming process. One of the challenges of gene editing is to produce transgene-free plants. The transgenes required to carry out selection and editing, such as antibiotic resistance markers and editing enzymes, must be removed, while the edited part of the genome is retained. This selection generally requires 1-3 generations of backcrossing, which takes time, particularly for long-generation species such as perennial plants. Huang et al. have addressed this problem by transiently expressing the gene-editing cassette. Because it is a transient expression, no foreign DNA will get incorporated into the genome. Specifically, the authors transiently expressed a base editor for targeted editing, a target gRNA construct, another gRNA that targets acetolactate synthase, and a GFP construct. While the target site will get a mutation, the other gRNA makes a mutation in acetolactate synthase leading to herbicide tolerance, which serves as a selection marker. Foreign DNA-containing plants show GFP expression, so this provides a negative screenable marker. Using this method, the authors were able to generate transgene-free gene-edited lines in the T0 generation of tomato, tobacco, potato, and citrus. As someone who performed transient assays throughout my PhD, I never imagined such a beautiful application of this technique. (Summary by Kamal Kumar Malukani, @KamalMalukani.) Nature Plants, 10.1038/s41477-023-01520-y.
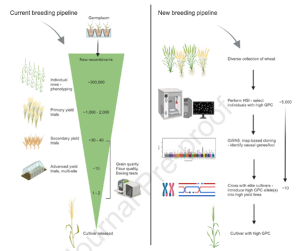
Genomic regions of durum wheat involved in water productivity
 Drought stress is a major problem and can cause large reductions in yield. Water productivity is the amount of yield per unit of water used, hence a higher water productivity is associated with more drought tolerance. Here, Zaïm et al. discovered three genomic loci associated with increased water productivity in durum wheat. They grew 384 durum wheat varieties in 18 environments which were classed as ‘moisture-stressed’ or ‘not moisture-stressed’. Using these environmental conditions and yield data, they conducted a genome wide association study (GWAS) and found six loci associated with yield under moisture-stress in multiple environments. To confirm this, the GWAS was repeated with a smaller panel of 80 different wheat varieties grown at two moisture-stressed locations, and three of the same loci, on chromosomes 1B, 2A and 7B, were discovered. The region on chromosome 1B contains a hydroxyproline-rich glycoprotein involved in protecting plants against water loss. The region on chromosome 2A contains transcription factors which could be involved in drought stress response, and the region on chromosome 7B contains genes encoding for aquaporins. All these loci have a positive effect on water productivity and thus positive alleles of these genes could be incorporated into durum wheat breeding programs. (Summary by Rose McNelly @Rose_McN) J Exp. Bot. 10.1093/jxb/erad357
Drought stress is a major problem and can cause large reductions in yield. Water productivity is the amount of yield per unit of water used, hence a higher water productivity is associated with more drought tolerance. Here, Zaïm et al. discovered three genomic loci associated with increased water productivity in durum wheat. They grew 384 durum wheat varieties in 18 environments which were classed as ‘moisture-stressed’ or ‘not moisture-stressed’. Using these environmental conditions and yield data, they conducted a genome wide association study (GWAS) and found six loci associated with yield under moisture-stress in multiple environments. To confirm this, the GWAS was repeated with a smaller panel of 80 different wheat varieties grown at two moisture-stressed locations, and three of the same loci, on chromosomes 1B, 2A and 7B, were discovered. The region on chromosome 1B contains a hydroxyproline-rich glycoprotein involved in protecting plants against water loss. The region on chromosome 2A contains transcription factors which could be involved in drought stress response, and the region on chromosome 7B contains genes encoding for aquaporins. All these loci have a positive effect on water productivity and thus positive alleles of these genes could be incorporated into durum wheat breeding programs. (Summary by Rose McNelly @Rose_McN) J Exp. Bot. 10.1093/jxb/erad357
Selective autophagy of THOUSAND GRAIN WEIGHT 6 protein
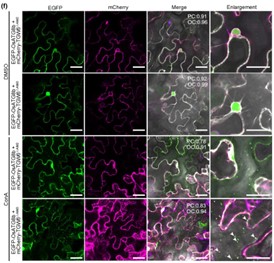 Rice is a globally important crop, therefore increasing yield without compromising quality is of much interest. A reduction in the amount of the TGW6 (THOUSAND-GRAIN WEIGHT 6) protein is associated with improved grain yield and quality in rice. However, we do not fully understand how TGW6 protein levels are controlled in developing rice grains. Here, bimolecular complementation and split luciferase assays were used to show that TGW6 interacts with the autophagosome component ATG8b in planta. This interaction was dependent on a motif called AIM2 in the TGW6 protein, as when the AIM2 motif was mutated the TGW6-ATG8b interaction was abolished. To test whether the interaction leads to autophagosome-induced degradation of TGW6, the authors transiently expressed fluorescent-tagged versions of TGW6, either with or without the AIM2 mutation, in Nicotiana benthamiana. Autophagic bodies were stabilized by application of Concanamycin A, allowing TGW6 puncta in autophagic bodies in the vacuole to be quantified. The authors found less TGW6 puncta when AIM2 was mutated, suggesting the TGW6-ATG8b interaction is needed for selective autophagy of TGW6. Hence, this study has unveiled a new mechanism by which TWG6 protein levels are controlled in plants and this knowledge could be used to further increase grain yield and quality in rice. (Summary by Rose McNelly @Rose_McN) New Phytol. 10.1111/nph.19271
Rice is a globally important crop, therefore increasing yield without compromising quality is of much interest. A reduction in the amount of the TGW6 (THOUSAND-GRAIN WEIGHT 6) protein is associated with improved grain yield and quality in rice. However, we do not fully understand how TGW6 protein levels are controlled in developing rice grains. Here, bimolecular complementation and split luciferase assays were used to show that TGW6 interacts with the autophagosome component ATG8b in planta. This interaction was dependent on a motif called AIM2 in the TGW6 protein, as when the AIM2 motif was mutated the TGW6-ATG8b interaction was abolished. To test whether the interaction leads to autophagosome-induced degradation of TGW6, the authors transiently expressed fluorescent-tagged versions of TGW6, either with or without the AIM2 mutation, in Nicotiana benthamiana. Autophagic bodies were stabilized by application of Concanamycin A, allowing TGW6 puncta in autophagic bodies in the vacuole to be quantified. The authors found less TGW6 puncta when AIM2 was mutated, suggesting the TGW6-ATG8b interaction is needed for selective autophagy of TGW6. Hence, this study has unveiled a new mechanism by which TWG6 protein levels are controlled in plants and this knowledge could be used to further increase grain yield and quality in rice. (Summary by Rose McNelly @Rose_McN) New Phytol. 10.1111/nph.19271
Many modes of Striga resistance in sorghum
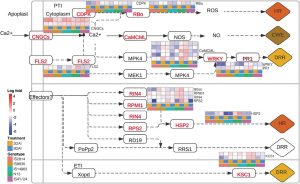 Witchweeds (Striga spp.) are parasitic plants. Like other weeds, they compete with food crops, but they do so very efficiently by penetrating host tissues and forming vascular connections. Through this effective extraction of nutrients and photosynthate from their hosts, Striga can literally wipe out yields in infected fields. Striga are most abundant in tropical regions including sub-Saharan Africa, and particularly affect sorghum, a staple crop in these regions. Here, Mutinda et al. used weighted gene co-expression network analyses (WGCNAs) to investigate how five Sorghum genotypes with some Striga resistance respond to infection. The lines analyzed, from diverse regions of Africa, likely acquired resistance independently, yet several common defense pathways were identified. Striga infection can activate pathogen-triggered immunity (PTI) and effector-triggered immunity (ETI) as well as salicylic and jasmonate-based defenses. Increased lignification to provide mechanical barriers and resist penetration also is a defense mechanism employed by some varieties. The WGCNA analysis further identified several interesting differentially regulated genes with known roles in defense [e.g., involved in callose synthesis, phosphoinositide signaling, and a thaumatin-like (PR-5) protein]. These results suggest several opportunities to breed or engineer sorghum with enhanced Striga resistance. (Summary by Mary Williams @PlantTeaching) J. Exp. Bot 10.1093/jxb/erad210
Witchweeds (Striga spp.) are parasitic plants. Like other weeds, they compete with food crops, but they do so very efficiently by penetrating host tissues and forming vascular connections. Through this effective extraction of nutrients and photosynthate from their hosts, Striga can literally wipe out yields in infected fields. Striga are most abundant in tropical regions including sub-Saharan Africa, and particularly affect sorghum, a staple crop in these regions. Here, Mutinda et al. used weighted gene co-expression network analyses (WGCNAs) to investigate how five Sorghum genotypes with some Striga resistance respond to infection. The lines analyzed, from diverse regions of Africa, likely acquired resistance independently, yet several common defense pathways were identified. Striga infection can activate pathogen-triggered immunity (PTI) and effector-triggered immunity (ETI) as well as salicylic and jasmonate-based defenses. Increased lignification to provide mechanical barriers and resist penetration also is a defense mechanism employed by some varieties. The WGCNA analysis further identified several interesting differentially regulated genes with known roles in defense [e.g., involved in callose synthesis, phosphoinositide signaling, and a thaumatin-like (PR-5) protein]. These results suggest several opportunities to breed or engineer sorghum with enhanced Striga resistance. (Summary by Mary Williams @PlantTeaching) J. Exp. Bot 10.1093/jxb/erad210
Evolution of research topics and paradigms in plant sciences
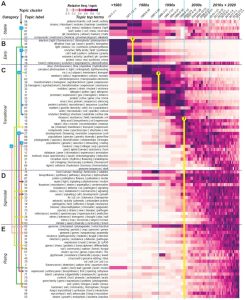 Is it possible to objectively summarize the recent history of plant science? Shiu and Lehti-Shiu have used a combination of machine learning and language modeling to track terms used in plant science articles from the 1950s to now, as a strategy to understand how research topics and tools have evolved. The records were clustered into 90 topics, which were named by the five most common terms defining them. As an example, the top topic since 1950 is listed as “QTL / resistance / wheat / markers / traits” and the third most common is “leaf / water / CO2 / trees / stomatal”. The topics were also clustered, forming for example a “development” topic cluster. Where this gets really interesting is where they start to look at trends over time. They identified five categories with different trends: stable, early, transitional, sigmoidal, and rising. Several of the rising categories include keywords that reflect current advancements in microbiome research, such as soil, rhizosphere, symbiosis, and mycorrhizal. Paradigm shifts from the pre-molecular era to the molecular era to the high-throughput -omics era are apparent (and thoughtfully discussed). The authors similarly discuss trends of taxa terms and research outputs by country. Interestingly, much of the growth in the past 20 years is attributed to research in India and China, and there has been an overall equalizing of plant science research with increasing impacts from countries without strong publication presences. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.02.560457
Is it possible to objectively summarize the recent history of plant science? Shiu and Lehti-Shiu have used a combination of machine learning and language modeling to track terms used in plant science articles from the 1950s to now, as a strategy to understand how research topics and tools have evolved. The records were clustered into 90 topics, which were named by the five most common terms defining them. As an example, the top topic since 1950 is listed as “QTL / resistance / wheat / markers / traits” and the third most common is “leaf / water / CO2 / trees / stomatal”. The topics were also clustered, forming for example a “development” topic cluster. Where this gets really interesting is where they start to look at trends over time. They identified five categories with different trends: stable, early, transitional, sigmoidal, and rising. Several of the rising categories include keywords that reflect current advancements in microbiome research, such as soil, rhizosphere, symbiosis, and mycorrhizal. Paradigm shifts from the pre-molecular era to the molecular era to the high-throughput -omics era are apparent (and thoughtfully discussed). The authors similarly discuss trends of taxa terms and research outputs by country. Interestingly, much of the growth in the past 20 years is attributed to research in India and China, and there has been an overall equalizing of plant science research with increasing impacts from countries without strong publication presences. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.02.560457