Plant Science Research Weekly: November 13, 2020
Review: Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution
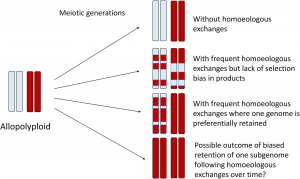 Polyploidy or whole-genome duplication (WGD) is an important process in plant evolution and speciation. Additional sets of chromosomes can be derived from intraspecific genome duplication (autopolyploidy) or hybridization of divergent genomes and chromosome doubling (allopolyploidy). In early stages of allopolyploid formation, the interaction and recombination between subgenomes (homoeologous exchange) is associated with changes in allele dosage, changes in methylation patterns, novel genomic structural variations and novel phenotypes. An additional category of polyploids has been described as “segmental allopolyploidy”, an intermediate point between autopolyploidy and allopolyploidy. This is, both recombination within subgenomes (disomic inheritance) and between subgenomes (tetrasomic inheritance) occur during meiosis. This generates a mosaic of genomic regions represented by both subgenomes, or by one or the other subgenome. In other words, in some genomic segments, one subgenome is deleted and replaced by segments of the other subgenome (not reciprocal exchange or biased replacement). In conclusion, homoeologous exchanges in allopolyploids is a driver of evolution, by generating evolutionary and phenotypic novelty. (Summary by Carolina Ballén-Taborda @carolinaballen) Front. Genetics 10.3389/fgene.2020.01014/full
Polyploidy or whole-genome duplication (WGD) is an important process in plant evolution and speciation. Additional sets of chromosomes can be derived from intraspecific genome duplication (autopolyploidy) or hybridization of divergent genomes and chromosome doubling (allopolyploidy). In early stages of allopolyploid formation, the interaction and recombination between subgenomes (homoeologous exchange) is associated with changes in allele dosage, changes in methylation patterns, novel genomic structural variations and novel phenotypes. An additional category of polyploids has been described as “segmental allopolyploidy”, an intermediate point between autopolyploidy and allopolyploidy. This is, both recombination within subgenomes (disomic inheritance) and between subgenomes (tetrasomic inheritance) occur during meiosis. This generates a mosaic of genomic regions represented by both subgenomes, or by one or the other subgenome. In other words, in some genomic segments, one subgenome is deleted and replaced by segments of the other subgenome (not reciprocal exchange or biased replacement). In conclusion, homoeologous exchanges in allopolyploids is a driver of evolution, by generating evolutionary and phenotypic novelty. (Summary by Carolina Ballén-Taborda @carolinaballen) Front. Genetics 10.3389/fgene.2020.01014/full
Gramene 2021: harnessing the power of comparative genomics and pathways for plant research
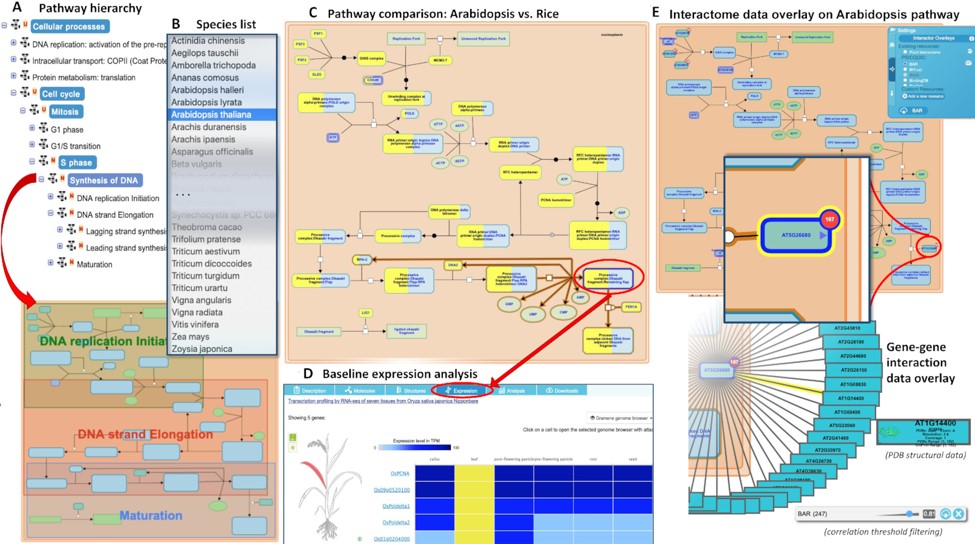
Gramene (www.gramene.org) is a curated, open-source, integrated data resource for comparative functional genomics in crops and model plant species. It launched 20 years ago and has continually grown and expanded to incorporate new tools and resources. This update by Tello-Ruiz et al. describes its current iteration, #63. Gramene “hosts 93 reference genomes—over 3.9 million genes in 122 947 families with orthologous and paralogous classifications.” As well as a Genome Browser, it includes an Expression Viewer and a Plant Reactome portal. As the authors point out, this extensive knowledgebase provides a “gold mine for training Machine Learning methods for knowledge extraction.” Through a pilot program called Domain Informational Vocabulary Extraction (DIVE), authors of recent Plant Cell and Plant Physiology papers have contributed to this effort by confirming or editing mined data. Future directions will include storing and providing access to species pan-genomes. (Summary by Mary Williams @PlantTeaching) Nucleic Acids Research 10.1093/nar/gkaa979
Condensation of Rubisco into a proto-pyrenoid in higher plant chloroplasts
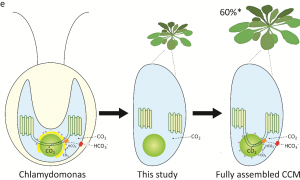 Rubisco is a major enzyme in assimilating CO2 during photosynthesis. The efficiency of CO2 assimilation is compromised by a low ratio of CO2/O2, especially in C3 plants like rice, wheat and soybean. Algae sequester Rubisco as a condensate in a microcompartment called a pyrenoid within the chloroplast and have a CO2-concentrating mechanism that delivers CO2 to the Rubisco within these sites. To address the feasibility of introducing a similar CO2-concentrating strategy to higher plants, Atkinson et al. expressed a mature EPYC1 (Essential Pyrenoid Component 1; a Rubisco-linking protein) and a plant-algal hybrid Rubisco in Arabidopsis chloroplasts. This expression led to Rubisco condensates, “proto-pyrenoids,” formation in the chloroplast. Further analysis of photosynthetic efficiency and biomass production of the transgenic plants showed no harmful effect. This study opens the possibility for increasing Rubisco CO2 assimilation efficiency in higher plants by next introducing the algal CO2 concentration mechanism to pump CO2 into the proto-pyrenoid. (Summary by Sunita Pathak @psunita980) bioRxiv 10.1101/2020.10.26.354332
Rubisco is a major enzyme in assimilating CO2 during photosynthesis. The efficiency of CO2 assimilation is compromised by a low ratio of CO2/O2, especially in C3 plants like rice, wheat and soybean. Algae sequester Rubisco as a condensate in a microcompartment called a pyrenoid within the chloroplast and have a CO2-concentrating mechanism that delivers CO2 to the Rubisco within these sites. To address the feasibility of introducing a similar CO2-concentrating strategy to higher plants, Atkinson et al. expressed a mature EPYC1 (Essential Pyrenoid Component 1; a Rubisco-linking protein) and a plant-algal hybrid Rubisco in Arabidopsis chloroplasts. This expression led to Rubisco condensates, “proto-pyrenoids,” formation in the chloroplast. Further analysis of photosynthetic efficiency and biomass production of the transgenic plants showed no harmful effect. This study opens the possibility for increasing Rubisco CO2 assimilation efficiency in higher plants by next introducing the algal CO2 concentration mechanism to pump CO2 into the proto-pyrenoid. (Summary by Sunita Pathak @psunita980) bioRxiv 10.1101/2020.10.26.354332
Conserved transcriptional programs underpin organogenesis and reproduction in land plants
 Land plant evolution accompanied a plethora of evolutionary novelties such as cells that compose complex organs (e.g., stems, leaves, roots) and male/female gametophytes. The identity and conservation of the transcriptional programs underlying organogenesis and reproductive development across land plants remains poorly explored. Julca et al., generated expression atlases of several organs and gametes for 10 representative species of major land plant lineages and performed comparative analyses to shed light on potentially conserved transcriptional signatures. They found that most organs, with the exception of roots and male gametes, express a significant part (>50%) of their genes, whereas roots and male gametes transcriptomes only express 46% and 38% of their genes, respectively, which suggests a higher level of specialization in these transcriptional programs. Phylostratigraphic and phylogenomic analyses showed that older gene families are more generally expressed (less organ-specific), whereas younger gene families tend to be more organ-specific, suggesting that new genes are more commonly recruited for specialized functions. In addition, many gene families that are expressed in specific organs appeared in the phylogeny long before the corresponding organs, which corroborates previous research: the establishment of new organs is strongly fueled by gene co-option. Analysis of male/female gametophyte transcriptomes revealed that transcriptional signatures of mature pollen are conserved across land plants, while female transcriptional programs are much less conserved. (Summary by Jesus Leon @jesussaur) bioRxiv 10.1101/2020.10.29.361501
Land plant evolution accompanied a plethora of evolutionary novelties such as cells that compose complex organs (e.g., stems, leaves, roots) and male/female gametophytes. The identity and conservation of the transcriptional programs underlying organogenesis and reproductive development across land plants remains poorly explored. Julca et al., generated expression atlases of several organs and gametes for 10 representative species of major land plant lineages and performed comparative analyses to shed light on potentially conserved transcriptional signatures. They found that most organs, with the exception of roots and male gametes, express a significant part (>50%) of their genes, whereas roots and male gametes transcriptomes only express 46% and 38% of their genes, respectively, which suggests a higher level of specialization in these transcriptional programs. Phylostratigraphic and phylogenomic analyses showed that older gene families are more generally expressed (less organ-specific), whereas younger gene families tend to be more organ-specific, suggesting that new genes are more commonly recruited for specialized functions. In addition, many gene families that are expressed in specific organs appeared in the phylogeny long before the corresponding organs, which corroborates previous research: the establishment of new organs is strongly fueled by gene co-option. Analysis of male/female gametophyte transcriptomes revealed that transcriptional signatures of mature pollen are conserved across land plants, while female transcriptional programs are much less conserved. (Summary by Jesus Leon @jesussaur) bioRxiv 10.1101/2020.10.29.361501
CAMEL in the canal: Regulation of PIN proteins during canalization
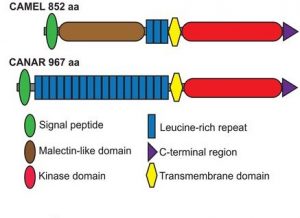 Auxin transport during development or wound regeneration is known to require auxin-induced channel or ‘canal’ formation that includes polar localization of PIN auxin transport proteins. Hajný and colleagues have characterized a receptor-like kinase (RLK) complex involved in auxin-induced pattern formation as well as PIN localization. The authors identified CAMEL, a membrane-localized RLK, and its interactor, CANAR, an auxin-responsive kinase, functioning downstream of the TIR1/AFB-WRKY23 transcriptional auxin signaling pathway. Knockout mutants of these proteins have disrupted auxin levels and abnormal leaf venation and wound regeneration responses, suggesting a direct role in auxin signaling or transport. Further biochemical and genetic investigations suggested CAMEL to be necessary for phosphorylation-dependent intracellular trafficking and localization of the PIN1 transporter. The authors propose CANAR to be a negative regulator of CAMEL. Thus, this study adds a novel component in the puzzle to solve auxin signaling and transport mechanisms. (Summary by Pavithran Narayanan @pavi_narayanan) Science 10.1126/science.aba3178
Auxin transport during development or wound regeneration is known to require auxin-induced channel or ‘canal’ formation that includes polar localization of PIN auxin transport proteins. Hajný and colleagues have characterized a receptor-like kinase (RLK) complex involved in auxin-induced pattern formation as well as PIN localization. The authors identified CAMEL, a membrane-localized RLK, and its interactor, CANAR, an auxin-responsive kinase, functioning downstream of the TIR1/AFB-WRKY23 transcriptional auxin signaling pathway. Knockout mutants of these proteins have disrupted auxin levels and abnormal leaf venation and wound regeneration responses, suggesting a direct role in auxin signaling or transport. Further biochemical and genetic investigations suggested CAMEL to be necessary for phosphorylation-dependent intracellular trafficking and localization of the PIN1 transporter. The authors propose CANAR to be a negative regulator of CAMEL. Thus, this study adds a novel component in the puzzle to solve auxin signaling and transport mechanisms. (Summary by Pavithran Narayanan @pavi_narayanan) Science 10.1126/science.aba3178
Formation of flavone-based wooly fibres by glandular trichomes of Dionysia tapetodes
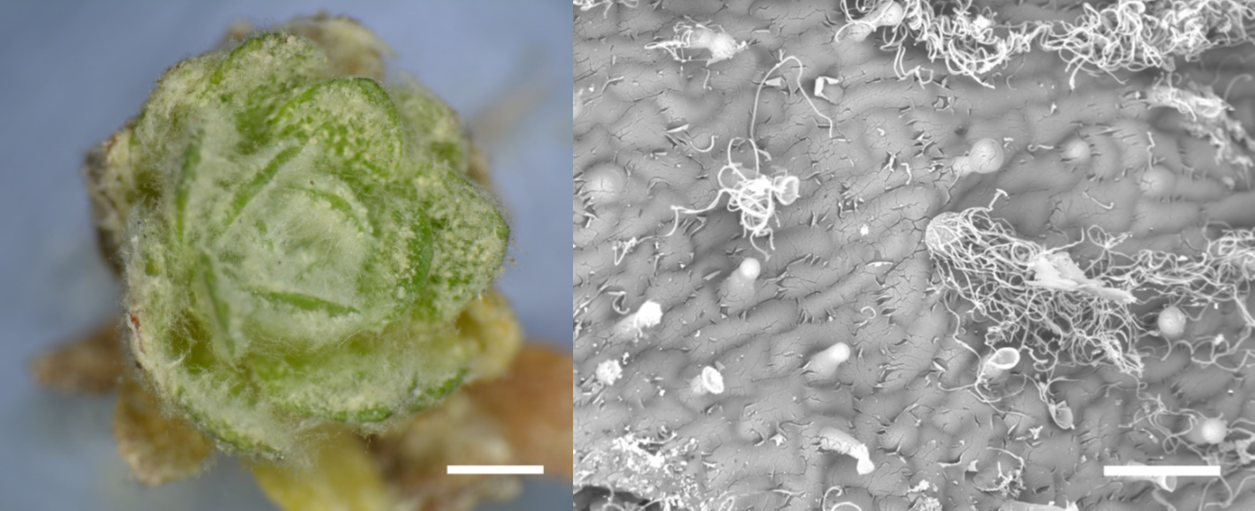 Many people are familiar with the popular houseplant Cephalocereus senilis, which also known as old man cactus because it is covered with long white hairs that are thought to protect it from frost and UV light. A similar function is described for the wooly fibers that cover the alpine plant Dionysia tapetodes, investigated here by Bourdon et al. The authors found that unlike related species that produce a powdery or dust-like covering or farina, D. tapetodes is covered with wool-like fibres averaging 1.6 um in diameter. They found that these fibres are extruded from a point in the heads of glandular trichomes, raising the questions of what they are made of and how are they assembled. Whilst powdery farina is made up of flavones, the fibres are made up of flavones and substituted flavones, which might provide the additional hydrogen bonding needed to maintain the fibre structure. TEM imaging suggests that the fibres are extruded through small holes in the glandular trichome heads from materials stored in the vacuole. As the first author noted on Twitter, these plants are able to “weave their own fabric”. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2020.10.06.320911
Many people are familiar with the popular houseplant Cephalocereus senilis, which also known as old man cactus because it is covered with long white hairs that are thought to protect it from frost and UV light. A similar function is described for the wooly fibers that cover the alpine plant Dionysia tapetodes, investigated here by Bourdon et al. The authors found that unlike related species that produce a powdery or dust-like covering or farina, D. tapetodes is covered with wool-like fibres averaging 1.6 um in diameter. They found that these fibres are extruded from a point in the heads of glandular trichomes, raising the questions of what they are made of and how are they assembled. Whilst powdery farina is made up of flavones, the fibres are made up of flavones and substituted flavones, which might provide the additional hydrogen bonding needed to maintain the fibre structure. TEM imaging suggests that the fibres are extruded through small holes in the glandular trichome heads from materials stored in the vacuole. As the first author noted on Twitter, these plants are able to “weave their own fabric”. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2020.10.06.320911
A novel family of secreted proteins linked to plant gall development
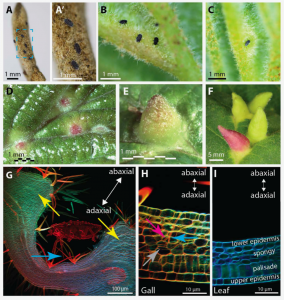 Insect-induced galls are abnormally patterned plant organs that provide insects with protection and a nutrient source, yet mechanisms underlying the development of these unique organs are mostly unknown. To identify insect factors that govern gall pattering, Korgaonkar et al. turned to the natural variation of the colors of galls induced by various insect species. They used the leaves of witch hazel, Hamamelis virginiana, as the host and the populations of aphid, Hormaphis cornu, as the galling insect model. A genome-wide association study on H. cornu isolated from 43 green galls and 47 red galls identified a gene, termed determinant of gall color (dgc), strongly associated with gall color; red galls were associated with weak dgc expression. Consistently, high levels of dgc transcription are associated with the downregulation of plant anthocyanin genes and two anthocyanins. Transcriptome analysis of H. cornu at different life cycles identified novel genes, termed bicycle genes (because of a conserved pair of CYC motifs; bi-CYC-like), which are specifically induced in the salivary glands of aphids only when they are able to induce galls. The authors propose that proteins encoded by bicycle genes, that are evolutionarily related to dgc and are under strong positive selection, may be secreted from gall-forming aphids into the host to affect various aspects of gall development. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.10.28.359562
Insect-induced galls are abnormally patterned plant organs that provide insects with protection and a nutrient source, yet mechanisms underlying the development of these unique organs are mostly unknown. To identify insect factors that govern gall pattering, Korgaonkar et al. turned to the natural variation of the colors of galls induced by various insect species. They used the leaves of witch hazel, Hamamelis virginiana, as the host and the populations of aphid, Hormaphis cornu, as the galling insect model. A genome-wide association study on H. cornu isolated from 43 green galls and 47 red galls identified a gene, termed determinant of gall color (dgc), strongly associated with gall color; red galls were associated with weak dgc expression. Consistently, high levels of dgc transcription are associated with the downregulation of plant anthocyanin genes and two anthocyanins. Transcriptome analysis of H. cornu at different life cycles identified novel genes, termed bicycle genes (because of a conserved pair of CYC motifs; bi-CYC-like), which are specifically induced in the salivary glands of aphids only when they are able to induce galls. The authors propose that proteins encoded by bicycle genes, that are evolutionarily related to dgc and are under strong positive selection, may be secreted from gall-forming aphids into the host to affect various aspects of gall development. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.10.28.359562
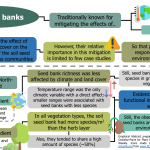
Buffering effects of soil seed banks on plant community composition in response to land use and climate
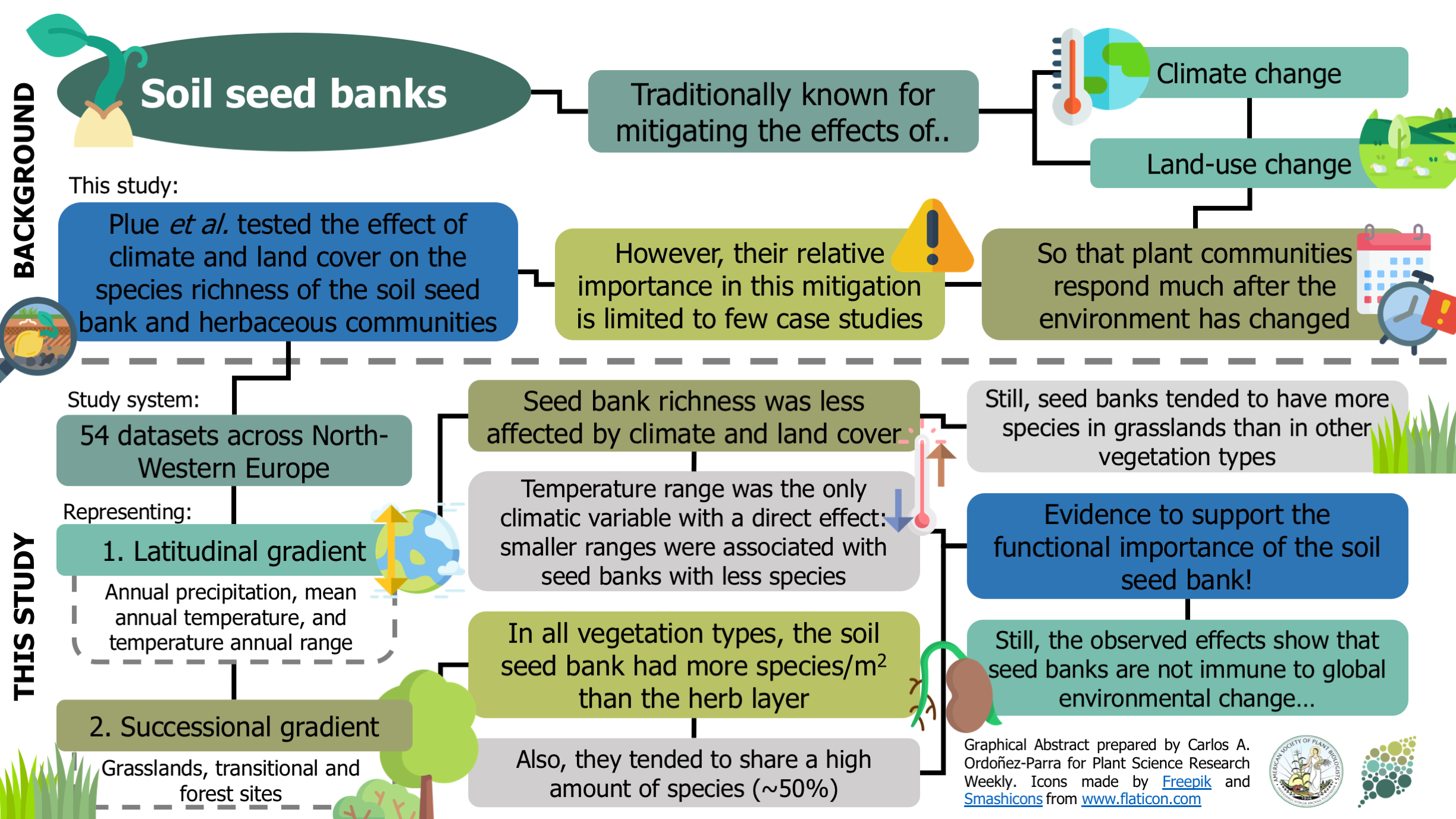
Plant communities seem to be able to mitigate the effects of climate and land-use change, given their responses are usually observed long after environmental conditions have changed. This lagged response has been attributed to the presence of persistent soil seed banks. Still, the evidence of their relative importance in this process is limited to individual case studies. Here, Plue and colleagues analyzed 54 datasets across a latitudinal and successional gradient in North-Western Europe to assess the effect of climate and vegetation type on soil seed banks and herb communities’ richness. Regardless of the vegetation type, the soil seed bank had more species per area than the herb layer and shared around half of its species. The richness of soil seed banks was less affected by environmental variables than the richness of standing communities, with annual temperature range the only climatic variable with significant direct effect. As a result, this research provides exciting insights that reinforce soil seed banks’ ecological importance in mitigating the effects of global environmental change on plant communities. However, the authors warn that this mitigating capacity of the soil seed banks could be compromised in the future due to increasing changes in the environment. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Glob. Ecol. Biogeogr. 10.1111/geb.13201