Plant Science Research Weekly: November 10, 2023
Review: Till death do us pair: Co-evolution of plant–necrotroph interactions
 This interesting and well-written review by Derbyshire and Raffaele takes a step back from the molecular interactions between plant and pathogen and discusses them in light of co-evolutionary processes. The review starts with a useful introduction and definition of concepts about “robustness” in biological systems, including multifunctionality, modularity, and compartmentalization, as well as the difference between degeneracy and robustness. The authors then apply these concepts to draw lessons about the co-evolutionary interactions between plants and necrotrophic pathogens, including that although programmed cell death is an effective strategy to combat biotrophic pathogens, it benefits necrotrophic pathogens, which obtain nutrients from dead tissues. I particularly enjoyed the discussion of the molecular factors that contribute to specialization versus generalism, such as the idea that heterogeneous host environments promote degeneracy in pathogen virulence genes. There’s also an interesting discussion about the spatial and cell-type specific interactions that are being illuminated by single-cell sequencing. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102457
This interesting and well-written review by Derbyshire and Raffaele takes a step back from the molecular interactions between plant and pathogen and discusses them in light of co-evolutionary processes. The review starts with a useful introduction and definition of concepts about “robustness” in biological systems, including multifunctionality, modularity, and compartmentalization, as well as the difference between degeneracy and robustness. The authors then apply these concepts to draw lessons about the co-evolutionary interactions between plants and necrotrophic pathogens, including that although programmed cell death is an effective strategy to combat biotrophic pathogens, it benefits necrotrophic pathogens, which obtain nutrients from dead tissues. I particularly enjoyed the discussion of the molecular factors that contribute to specialization versus generalism, such as the idea that heterogeneous host environments promote degeneracy in pathogen virulence genes. There’s also an interesting discussion about the spatial and cell-type specific interactions that are being illuminated by single-cell sequencing. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2023.102457
Spotlight: Super-pangenomes for improved breeding
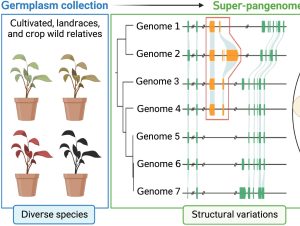 Sometimes more really is better, and I think it’s safe to say that when it comes to genomic information, more is better. Here, Raza et al. highlight the great value of super-pangenomes. A pan-genome is defined as the entire set of genes within a species, created by combining sequences of many individuals. The super-pangenome as defined here expands this concept to incorporate genomes of close relatives, up to the genus level. This idea is particularly useful as a way to incorporate traits from crop wild relatives into breeding programs. Crop wild relatives often are sources of genes the provide environmental robustness that has been lost through selection for yields, but introgressions of these valuable traits can be difficult because of structural variations that have arisen over time. The authors cite examples of super-pangenome assemblies that have been developed for rice, soybean, and tomato, and how these resources have allowed the identification of several novel genes. Several strategies for breeding using information from super-pangenomes is also discussed. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.08.005
Sometimes more really is better, and I think it’s safe to say that when it comes to genomic information, more is better. Here, Raza et al. highlight the great value of super-pangenomes. A pan-genome is defined as the entire set of genes within a species, created by combining sequences of many individuals. The super-pangenome as defined here expands this concept to incorporate genomes of close relatives, up to the genus level. This idea is particularly useful as a way to incorporate traits from crop wild relatives into breeding programs. Crop wild relatives often are sources of genes the provide environmental robustness that has been lost through selection for yields, but introgressions of these valuable traits can be difficult because of structural variations that have arisen over time. The authors cite examples of super-pangenome assemblies that have been developed for rice, soybean, and tomato, and how these resources have allowed the identification of several novel genes. Several strategies for breeding using information from super-pangenomes is also discussed. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.08.005
Chloroplast proteostasis prevents aggregation of Huntington’s disease-causing human polyQ protein
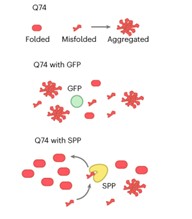 Certain human neurodegenerative disorders are caused by aggregation of disordered proteins. In particular, Huntington’s disease is caused by aggregation of a protein called huntingtin, which contains long stretches of glutamine (Q). Llama et al. observed that proteins with long stretches of glutamine are also present in plants, but they do not seem to cause disease. To investigate how plants control aggregation of these polyQ-containing proteins, the authors generated transgenic Arabidopsis thaliana expressing a fragment of human huntingtin labelled with a fluorophore. Using confocal microscopy, they observed that huntingtin remains soluble in plant cells in normal growth conditions but becomes aggregated under heat stress. They found that the expressed protein interacts with components of chloroplast protein import and chloroplast proteases, and that impairment of these chloroplast functions caused aggregation of huntingtin in plant cells. Thus, the authors concluded that plants use chloroplast proteostasis to control aggregation of polyglutamine proteins. Expression of a chloroplast protease, the chloroplast stromal processing peptidase (SPP), can also prevent aggregation of huntingtin both in a human cell line and in nematodes. This work presents plant chloroplast proteostasis as a novel path towards developing treatments for human neurodegenerative diseases. (Summary by Ángel Vergara @ngelVerCru) ) Nature Aging 10.1038/s43587-023-00502-1
Certain human neurodegenerative disorders are caused by aggregation of disordered proteins. In particular, Huntington’s disease is caused by aggregation of a protein called huntingtin, which contains long stretches of glutamine (Q). Llama et al. observed that proteins with long stretches of glutamine are also present in plants, but they do not seem to cause disease. To investigate how plants control aggregation of these polyQ-containing proteins, the authors generated transgenic Arabidopsis thaliana expressing a fragment of human huntingtin labelled with a fluorophore. Using confocal microscopy, they observed that huntingtin remains soluble in plant cells in normal growth conditions but becomes aggregated under heat stress. They found that the expressed protein interacts with components of chloroplast protein import and chloroplast proteases, and that impairment of these chloroplast functions caused aggregation of huntingtin in plant cells. Thus, the authors concluded that plants use chloroplast proteostasis to control aggregation of polyglutamine proteins. Expression of a chloroplast protease, the chloroplast stromal processing peptidase (SPP), can also prevent aggregation of huntingtin both in a human cell line and in nematodes. This work presents plant chloroplast proteostasis as a novel path towards developing treatments for human neurodegenerative diseases. (Summary by Ángel Vergara @ngelVerCru) ) Nature Aging 10.1038/s43587-023-00502-1
Altering chloroplast biogenesis leads to increased yields in rice
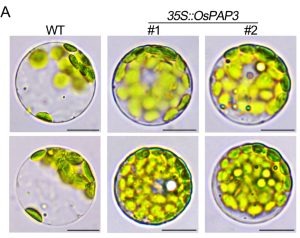 Transcription of chloroplast genes is carried out by the plastid encoded polymerase (PEP) with help from PAPs (PEP-associated proteins). PAP3 is important for chloroplast development in Arabidopsis thaliana and Nicotiana benthamiana, however its role in crops has not been fully elucidated. Here Seo et al. transformed rice to express OsPAP3 under the constitutively active 35S promoter. These plants had a 40-fold increase in PAP3 expression and were greener due to a significant increase in chlorophyll content. To assess the effect of PAP3 overexpression on chloroplast development, they quantified chloroplast number in one-week old protoplasts. For wild type plants there were approximately 14 chloroplasts per protoplast, whilst for the overexpression lines chloroplast number increased and there was an average of 21 chloroplasts per protoplast. To see if this caused an increase in plant productivity, the overexpression lines were grown in a paddy field for three consecutive years, and it was found that these plants had a 18-30% increase in yield. Hence altering chloroplast biogenesis could be an effective strategy to increase yields in crops. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad536
Transcription of chloroplast genes is carried out by the plastid encoded polymerase (PEP) with help from PAPs (PEP-associated proteins). PAP3 is important for chloroplast development in Arabidopsis thaliana and Nicotiana benthamiana, however its role in crops has not been fully elucidated. Here Seo et al. transformed rice to express OsPAP3 under the constitutively active 35S promoter. These plants had a 40-fold increase in PAP3 expression and were greener due to a significant increase in chlorophyll content. To assess the effect of PAP3 overexpression on chloroplast development, they quantified chloroplast number in one-week old protoplasts. For wild type plants there were approximately 14 chloroplasts per protoplast, whilst for the overexpression lines chloroplast number increased and there was an average of 21 chloroplasts per protoplast. To see if this caused an increase in plant productivity, the overexpression lines were grown in a paddy field for three consecutive years, and it was found that these plants had a 18-30% increase in yield. Hence altering chloroplast biogenesis could be an effective strategy to increase yields in crops. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad536
Staining starch granules in living plants
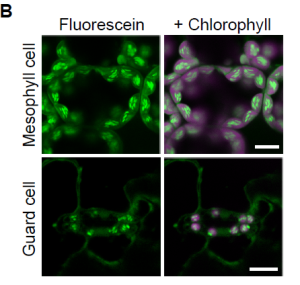 Starch granules are synthesized in the leaves of chloroplast during the day and degraded in the subsequent night. To visualise starch granules the tissue usually has to be fixed, which prevents live imaging. Here Ichikawa et al. set out to find a highly specific dye to image starch granules in living tissues. They tested a range of fluorescent dyes on Arabidopsis thaliana leaves and found that fluorescein stained granular structures in the mesophyll, guard cells and root tip. To test whether these were starch granules, they treated plants expressing the genetically encoded starch granule marker GBSSI-tagRFP (GRANULE BOUND STARCH SYNTHASE-RED FLUORESCENT PROTEIN) with fluorescein. The fluorescein signal overlapped with the RFP signal, confirming that fluorescein stains starch granules. The authors tested whether fluorescein could be used in other species and found it successfully stained starch granules in the bryophyte Marchantia polymorpha, and other flowering plants including strawberry, tomato and lettuce. Hence fluorescein provides an easy way to image starch granules in different species. (Summary by Rose McNelly @Rose_McN) Plant Physiol.10.1093/plphys/kiad528
Starch granules are synthesized in the leaves of chloroplast during the day and degraded in the subsequent night. To visualise starch granules the tissue usually has to be fixed, which prevents live imaging. Here Ichikawa et al. set out to find a highly specific dye to image starch granules in living tissues. They tested a range of fluorescent dyes on Arabidopsis thaliana leaves and found that fluorescein stained granular structures in the mesophyll, guard cells and root tip. To test whether these were starch granules, they treated plants expressing the genetically encoded starch granule marker GBSSI-tagRFP (GRANULE BOUND STARCH SYNTHASE-RED FLUORESCENT PROTEIN) with fluorescein. The fluorescein signal overlapped with the RFP signal, confirming that fluorescein stains starch granules. The authors tested whether fluorescein could be used in other species and found it successfully stained starch granules in the bryophyte Marchantia polymorpha, and other flowering plants including strawberry, tomato and lettuce. Hence fluorescein provides an easy way to image starch granules in different species. (Summary by Rose McNelly @Rose_McN) Plant Physiol.10.1093/plphys/kiad528
When and how did carrots turn orange?
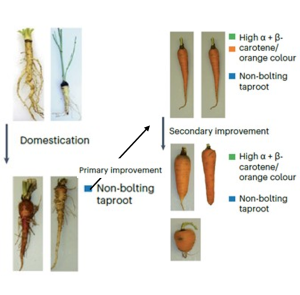 Carrots were not always orange, and a new paper by Coe, Bostan, Rolling et al. sheds new light into the history of carrot domestication and improvement, i.e., how we went from white, knotty carrots to the orange, smooth ones that are now consumed all over the world. The authors published a new version of the carrot genome that covers over 90% of its total estimated size, and they analyzed over 600 carrot accessions to reveal that carrot domestication first started in central Asia about 1200 years ago, followed by selection for orange carrots around 500 years ago. Contrary to previous reports, the authors propose that domestication and improvement resulted in a decrease in genetic diversity in cultivated carrot accessions. Carrot domestication and improvement involved the selection of crucial developmental traits including delayed flowering time, as the taproot becomes inedible upon flowering. The authors identified a gene in each of three crucial quantitative trait loci associated with the orange phenotype of modern carrots, involved in carotenoid biosynthesis, light perception responses and chloroplast biogenesis. Recessive alleles of these three genes are responsible for α-carotene and β-carotene accumulation, which gives the striking orange phenotype. These findings are in line with historical documents related to carrot domestication and improvement, and they provide new insights into the molecular mechanisms underlying the familiar orange phenotype. (Summary by Laura Turchi @turchi_l) Nature Plants 10.1038/s41477-023-01526-6
Carrots were not always orange, and a new paper by Coe, Bostan, Rolling et al. sheds new light into the history of carrot domestication and improvement, i.e., how we went from white, knotty carrots to the orange, smooth ones that are now consumed all over the world. The authors published a new version of the carrot genome that covers over 90% of its total estimated size, and they analyzed over 600 carrot accessions to reveal that carrot domestication first started in central Asia about 1200 years ago, followed by selection for orange carrots around 500 years ago. Contrary to previous reports, the authors propose that domestication and improvement resulted in a decrease in genetic diversity in cultivated carrot accessions. Carrot domestication and improvement involved the selection of crucial developmental traits including delayed flowering time, as the taproot becomes inedible upon flowering. The authors identified a gene in each of three crucial quantitative trait loci associated with the orange phenotype of modern carrots, involved in carotenoid biosynthesis, light perception responses and chloroplast biogenesis. Recessive alleles of these three genes are responsible for α-carotene and β-carotene accumulation, which gives the striking orange phenotype. These findings are in line with historical documents related to carrot domestication and improvement, and they provide new insights into the molecular mechanisms underlying the familiar orange phenotype. (Summary by Laura Turchi @turchi_l) Nature Plants 10.1038/s41477-023-01526-6
Physcomitrium patens responses to elevated CO2 and nitrogen
 With atmospheric CO2 levels increasing 100 times faster than historical levels, much attention has been paid to how crop plants respond to this elevated CO2 (eCO2). For angiosperms, eCO2 leads to increased CO2 assimilation and decreased photorespiration, but over time plants can adapt, leading to lower stomatal conductivity, and ultimately lower rates of nitrogen assimilation. Although not significant food sources for humans, mosses are important carbon sinks and critical components of many ecosystems, yet their responses to eCO2 have not been examined in detail. Here, Mohanasundaram et al. explored how the model moss Physcomitrium patens responds to eCO2. The authors measured growth rates and growth patterns and quantified key metabolites. They noted faster growth and life cycle progression under eCO2, with similar effects upon glucose treatment; these are likely responses to enhanced sugar signaling. The growth patterns were also impacted by the availability of nitrogen in the growth medium; for example, elevated nitrogen preferentially promoted growth in photosynthetic tissues. These findings demonstrate coherent moss responses to changes in exogenous CO2 and nitrogen levels, which can be useful for future ecosystem and climate models. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19348
With atmospheric CO2 levels increasing 100 times faster than historical levels, much attention has been paid to how crop plants respond to this elevated CO2 (eCO2). For angiosperms, eCO2 leads to increased CO2 assimilation and decreased photorespiration, but over time plants can adapt, leading to lower stomatal conductivity, and ultimately lower rates of nitrogen assimilation. Although not significant food sources for humans, mosses are important carbon sinks and critical components of many ecosystems, yet their responses to eCO2 have not been examined in detail. Here, Mohanasundaram et al. explored how the model moss Physcomitrium patens responds to eCO2. The authors measured growth rates and growth patterns and quantified key metabolites. They noted faster growth and life cycle progression under eCO2, with similar effects upon glucose treatment; these are likely responses to enhanced sugar signaling. The growth patterns were also impacted by the availability of nitrogen in the growth medium; for example, elevated nitrogen preferentially promoted growth in photosynthetic tissues. These findings demonstrate coherent moss responses to changes in exogenous CO2 and nitrogen levels, which can be useful for future ecosystem and climate models. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19348
Single-nucleus sequencing reveals transition from C3 to C4 photosynthesis
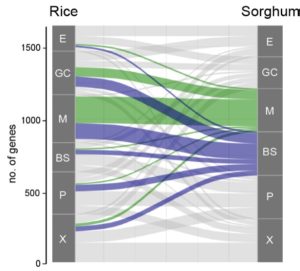 Sometime in the Cretaceous period (dinosaur time!), some monocots acquired a special pathway for carbon fixation, rendering them more efficient particularly in hot or dry environments. In most monocots, carbon is fixed by Rubisco in the mesophyll cells. In the innovative pathway, carbon fixation is split into two cell types, with the initial capture of CO2 taking place in the mesophyll cells by the enzyme PEPC to form a four-carbon product which delivers substrate to Rubisco, localized in the bundle sheath cell; this pathway is called C4 to reflect the novel four-carbon intermediate. To understand the origin of the C4 pathway, Swift and Luginbuehl et al. conducted a transcriptome comparison between rice (C3) and sorghum (C4) plants, with a particular focus on mesophyll and bundle sheath cells. The authors started by RNA sequencing of isolated nuclei, eliminating the problems associated with tissue-level transcriptomics and providing increased granularity. One of the main results of this elegant work is that genes that are normally expressed in mesophyll cells in rice are now active in the bundle sheath cells of sorghum, accomplished by the genes acquiring the relevant transcription factor binding sites (cis-elements). These data not only provide a glimpse into the evolution of C4 photosynthesis, but provide important clues for the efforts to engineer the more efficient C4 pathway into C3 plants such as rice. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.26.562893
Sometime in the Cretaceous period (dinosaur time!), some monocots acquired a special pathway for carbon fixation, rendering them more efficient particularly in hot or dry environments. In most monocots, carbon is fixed by Rubisco in the mesophyll cells. In the innovative pathway, carbon fixation is split into two cell types, with the initial capture of CO2 taking place in the mesophyll cells by the enzyme PEPC to form a four-carbon product which delivers substrate to Rubisco, localized in the bundle sheath cell; this pathway is called C4 to reflect the novel four-carbon intermediate. To understand the origin of the C4 pathway, Swift and Luginbuehl et al. conducted a transcriptome comparison between rice (C3) and sorghum (C4) plants, with a particular focus on mesophyll and bundle sheath cells. The authors started by RNA sequencing of isolated nuclei, eliminating the problems associated with tissue-level transcriptomics and providing increased granularity. One of the main results of this elegant work is that genes that are normally expressed in mesophyll cells in rice are now active in the bundle sheath cells of sorghum, accomplished by the genes acquiring the relevant transcription factor binding sites (cis-elements). These data not only provide a glimpse into the evolution of C4 photosynthesis, but provide important clues for the efforts to engineer the more efficient C4 pathway into C3 plants such as rice. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.26.562893
A dirigent protein complex directs lignin polymerization and assembly of the root diffusion barrier
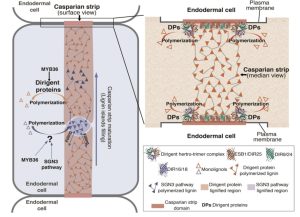 In Latin, dirigere means “to align or guide”, so in 1997 when a protein was identified that guides the stereoselectivity of another enzyme it was named as a dirigent protein. Subsequent studies found that the Arabidopsis genome encodes 25 dirigent proteins (DPs). In this new work, Gao et al. characterized the expression and function of several of these DPs. The authors first noted that five DPs are expressed preferentially in the root endodermal cell layer. Fluorescent protein fusions showed that they are localized at the Casparian strip, a lignified barrier that controls movement into the central part of the root. Various single- and double-mutant knock outs showed defects in Casparian strip lignification and function and mislocalization of the CASP-1 protein. Characteristically, at the Casparian strip the plasma membrane is tightly adhered to the cell wall, and this is also disrupted in DP mutants. The original dirigent protein was identified by its influence on lignin biosynthesis, and here the authors also found that the endodermal DPs are required for proper polymerization of monolignols and deposition of lignin in the early stages of Casparian strip formation. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adi503
In Latin, dirigere means “to align or guide”, so in 1997 when a protein was identified that guides the stereoselectivity of another enzyme it was named as a dirigent protein. Subsequent studies found that the Arabidopsis genome encodes 25 dirigent proteins (DPs). In this new work, Gao et al. characterized the expression and function of several of these DPs. The authors first noted that five DPs are expressed preferentially in the root endodermal cell layer. Fluorescent protein fusions showed that they are localized at the Casparian strip, a lignified barrier that controls movement into the central part of the root. Various single- and double-mutant knock outs showed defects in Casparian strip lignification and function and mislocalization of the CASP-1 protein. Characteristically, at the Casparian strip the plasma membrane is tightly adhered to the cell wall, and this is also disrupted in DP mutants. The original dirigent protein was identified by its influence on lignin biosynthesis, and here the authors also found that the endodermal DPs are required for proper polymerization of monolignols and deposition of lignin in the early stages of Casparian strip formation. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adi503
Bacterial tolerance to host-exuded specialized metabolites structures the maize root microbiome
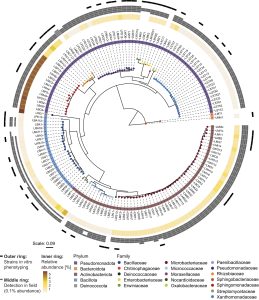 Benzoxazinoids (BXs) are alkaloid specialized metabolites produced by important crops like maize. Their role in shaping the root and rhizosphere microbiomes and in plant defense against pest and pathogens is well known, although the mechanism remains unknown. One of the major BXs produced by maize is DIMBOA-Glc, which is rapidly converted to MBOA in the rhizosphere and can suppress growth of many bacteria. Thoenen et al. found that these plant metabolites act as highly selective anti-microbials, with gram-positive bacteria such as Bacillaceae being more tolerant to MBOA than gram-negative bacteria (Xanthomonadaceae and Rhizobiaceae), revealing that cell wall properties are important for bacterial tolerance to BXs. The authors further examined responses of bacteria isolated from both host (maize) and non-host (Arabidopsis) plants, and found that even within the same taxonomic group, the maize root bacteria were more tolerant to MBOA. The authors systematically determined the tolerances of maize root bacteria to BXs and found that tolerance to MBOA correlated significantly with their abundance on BX-exuding roots. This study provides evidence that bacterial tolerance to plant specialized metabolites or to root-derived microbials is key to understanding characteristic root microbiome composition and its abundance in host plants. (Summary by Indrani Kakati, @Indranik333) PNAS 10.1073/pnas.2310134120
Benzoxazinoids (BXs) are alkaloid specialized metabolites produced by important crops like maize. Their role in shaping the root and rhizosphere microbiomes and in plant defense against pest and pathogens is well known, although the mechanism remains unknown. One of the major BXs produced by maize is DIMBOA-Glc, which is rapidly converted to MBOA in the rhizosphere and can suppress growth of many bacteria. Thoenen et al. found that these plant metabolites act as highly selective anti-microbials, with gram-positive bacteria such as Bacillaceae being more tolerant to MBOA than gram-negative bacteria (Xanthomonadaceae and Rhizobiaceae), revealing that cell wall properties are important for bacterial tolerance to BXs. The authors further examined responses of bacteria isolated from both host (maize) and non-host (Arabidopsis) plants, and found that even within the same taxonomic group, the maize root bacteria were more tolerant to MBOA. The authors systematically determined the tolerances of maize root bacteria to BXs and found that tolerance to MBOA correlated significantly with their abundance on BX-exuding roots. This study provides evidence that bacterial tolerance to plant specialized metabolites or to root-derived microbials is key to understanding characteristic root microbiome composition and its abundance in host plants. (Summary by Indrani Kakati, @Indranik333) PNAS 10.1073/pnas.2310134120
Building an inclusive botany: A radical dream
 Recent years have seen a reevaluation of the history and practice of science, including reconsidering who is recognized and why. Science doesn’t exist in a vacuum; it reflects the culture and values of those that practice it. This article, by Mabry et al., is intended to serve as a conversation starter to evaluate scientific botany in light of historical (and ongoing) exclusionary and exploitative practices. The article begins by highlighting the exceptions: plants with names that honor women or Latine, Black, or Indigenous people, and people who against the odds were otherwise recognized for their contributions to botany. The article also discusses plants with problematic names that refer to racist or exclusionary practices. Finally, the authors look forward to opportunities to move towards a more inclusive botany and recommend practices to get there, with a particular focus on specimen digitization. Digital collections are more accessible than those held solely as physical objects, and they also provide opportunities to include additional names (both taxonomic synonyms and Indigenous names) and to recognize the contributions from Traditional Knowledge (for example through Biocultural Labels https://localcontexts.org/). This article will soon be accompanied by learning materials – watch this space! (Summary by Mary Williams @PlantTeaching) ecoevoRxiv https://doi.org/10.32942/X22P51
Recent years have seen a reevaluation of the history and practice of science, including reconsidering who is recognized and why. Science doesn’t exist in a vacuum; it reflects the culture and values of those that practice it. This article, by Mabry et al., is intended to serve as a conversation starter to evaluate scientific botany in light of historical (and ongoing) exclusionary and exploitative practices. The article begins by highlighting the exceptions: plants with names that honor women or Latine, Black, or Indigenous people, and people who against the odds were otherwise recognized for their contributions to botany. The article also discusses plants with problematic names that refer to racist or exclusionary practices. Finally, the authors look forward to opportunities to move towards a more inclusive botany and recommend practices to get there, with a particular focus on specimen digitization. Digital collections are more accessible than those held solely as physical objects, and they also provide opportunities to include additional names (both taxonomic synonyms and Indigenous names) and to recognize the contributions from Traditional Knowledge (for example through Biocultural Labels https://localcontexts.org/). This article will soon be accompanied by learning materials – watch this space! (Summary by Mary Williams @PlantTeaching) ecoevoRxiv https://doi.org/10.32942/X22P51



