Plant Science Research Weekly: May 31st
Review: Integration of sulfate assimilation with C and N metabolism in transition from C3 to C4 photosynthesis
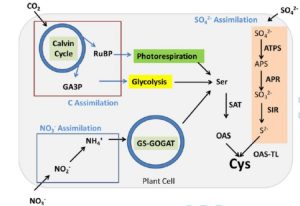 “Cysteine (HO2CCH(NH2)CH2SH) synthesis is the converging point of the three major pathways of primary metabolism: carbon, nitrate, and sulfate assimilation.” These metabolic connections are revealed in that a deficiency in one nutrient affects assimilation rates of the others. As yet, the connections between pathways are still being discovered, but in plants, the cysteine precursor O-acetylserine (OAS) and sulfide seem to be sensed. The coordination between C, N and S is rendered more complex in C4 plants, in which aspects of their assimilation is distributed between mesophyll and bundle sheath cells. In this review, Jobe et al. summarize the knowns and unknowns about how these three pathways are coordinated. They observe that efforts to engineer more-efficient C4 photosynthesis into C3 plants (like rice) will require a greater understanding of how C4 metabolism affects N and S assimilation. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz250
“Cysteine (HO2CCH(NH2)CH2SH) synthesis is the converging point of the three major pathways of primary metabolism: carbon, nitrate, and sulfate assimilation.” These metabolic connections are revealed in that a deficiency in one nutrient affects assimilation rates of the others. As yet, the connections between pathways are still being discovered, but in plants, the cysteine precursor O-acetylserine (OAS) and sulfide seem to be sensed. The coordination between C, N and S is rendered more complex in C4 plants, in which aspects of their assimilation is distributed between mesophyll and bundle sheath cells. In this review, Jobe et al. summarize the knowns and unknowns about how these three pathways are coordinated. They observe that efforts to engineer more-efficient C4 photosynthesis into C3 plants (like rice) will require a greater understanding of how C4 metabolism affects N and S assimilation. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz250
Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock ($)
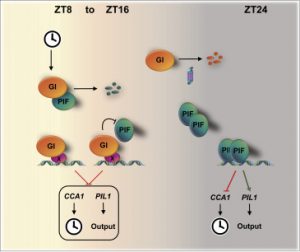 The daily cycle of light and dark controls many facets of plant growth and development, but these cycles depend on and are most efficient when integrated with the rhythms of the circadian clock. Nohales et al. have identified a key mechanism that directly links the regulation of light-responsive genes and the regulation of the clock. GIGANTEA (GI) is a plant-specific protein that has been shown genetically to contribute to light and clock signaling but whose biochemical function has been difficult to determine. Here, the authors show that GI binds to the promoter regions of numerous genes, perhaps indirectly through interactions with other transcription factors. The authors also show that GI interacts with PIF proteins, modulating their stability and activity. Finally, they show that “modulation of PIF activity by GI is not only required to adequately phase output rhythms such as growth but also for proper clock progression and light input.” (Summary by Mary Williams) Devel. Cell 10.1016/j.devcel.2019.04.030
The daily cycle of light and dark controls many facets of plant growth and development, but these cycles depend on and are most efficient when integrated with the rhythms of the circadian clock. Nohales et al. have identified a key mechanism that directly links the regulation of light-responsive genes and the regulation of the clock. GIGANTEA (GI) is a plant-specific protein that has been shown genetically to contribute to light and clock signaling but whose biochemical function has been difficult to determine. Here, the authors show that GI binds to the promoter regions of numerous genes, perhaps indirectly through interactions with other transcription factors. The authors also show that GI interacts with PIF proteins, modulating their stability and activity. Finally, they show that “modulation of PIF activity by GI is not only required to adequately phase output rhythms such as growth but also for proper clock progression and light input.” (Summary by Mary Williams) Devel. Cell 10.1016/j.devcel.2019.04.030
Accelerated flowering time reduces lifetime water use without penalizing reproductive performance in Arabidopsis
 As climate change proceeds, the availability of fresh water is decreasing. Luckily, the variation in water use efficiency within and between species provides tools necessary to breed plants for future climate. However, water use is often studied at the vegetative level. Ferguson et al. observed that whole plant water use and vegetative water use efficiency have different genetic makeup in mapping populations of Arabidopsis. The “more crop per drop” phenotype was associated with non-functional alleles of Flowering Locus C (FLC) and FRIGIDA due to their effects on flowering time. Interestingly, the reduced lifetime use of water did not penalize plant reproductive fitness. This study suggests that focusing solely on water use efficiency during the vegetative stage might not be indicative of crop productivity. (Summary by Magdalena Julkowska) Plant Cell Environ. 10.1111/pce.13527
As climate change proceeds, the availability of fresh water is decreasing. Luckily, the variation in water use efficiency within and between species provides tools necessary to breed plants for future climate. However, water use is often studied at the vegetative level. Ferguson et al. observed that whole plant water use and vegetative water use efficiency have different genetic makeup in mapping populations of Arabidopsis. The “more crop per drop” phenotype was associated with non-functional alleles of Flowering Locus C (FLC) and FRIGIDA due to their effects on flowering time. Interestingly, the reduced lifetime use of water did not penalize plant reproductive fitness. This study suggests that focusing solely on water use efficiency during the vegetative stage might not be indicative of crop productivity. (Summary by Magdalena Julkowska) Plant Cell Environ. 10.1111/pce.13527
Three-dimensional time-lapse analysis of maize root system archictecture
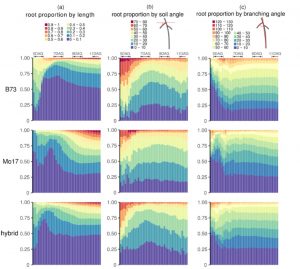 Root system architecture (RSA) profoundly affects plant nutritent uptake and response to drought, and is also famously extremely developmental plastic, which makes it difficult to identify genes that control root growth traits. Here, Jiang et al. analyzed 3D growth patterns over time of three maize lines (Mo17, B73 and their hybrid) growing in a gel-based growth medium. From these images, they extracted structural and dynamic traits (e.g., total root length and volume, and growth rate and orientation for each individual root branch). They then developed mathematical models to describe the RSA dynamics for each line. They found clear genotype-specific differences in global RSAs over time. Furthemore, these differences were also apparent when they compared data from gel-grown plants to data from field-grown plants, indicating that this approach could, “eventually be used to predict genetic variation for complex RSAs and their functions.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00015
Root system architecture (RSA) profoundly affects plant nutritent uptake and response to drought, and is also famously extremely developmental plastic, which makes it difficult to identify genes that control root growth traits. Here, Jiang et al. analyzed 3D growth patterns over time of three maize lines (Mo17, B73 and their hybrid) growing in a gel-based growth medium. From these images, they extracted structural and dynamic traits (e.g., total root length and volume, and growth rate and orientation for each individual root branch). They then developed mathematical models to describe the RSA dynamics for each line. They found clear genotype-specific differences in global RSAs over time. Furthemore, these differences were also apparent when they compared data from gel-grown plants to data from field-grown plants, indicating that this approach could, “eventually be used to predict genetic variation for complex RSAs and their functions.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00015
GRANAR, a new computational tool to better understand the functional importance of root anatomy
 Uptake of water by plants depends on root conductivity, which in turn is determined by hydraulic properties of individual cells and cell anatomy. Quantification of radial root anatomy is a time-consuming process, limiting our understanding of root anatomical features contributing to water uptake. Heymans et al. developed a computational tool Generator of Root ANAtomy in R (GRANAR), which allows for accelerated digitalization of radial root anatomy. The authors successfully re-analyzed maize root anatomies, and by combining GRANAR with MECHA, the hydraulic properties of individual roots could be estimated. GRANAR also allowed predicting conductivity of theoretical root anatomies, revealing the importance of individual root anatomy features, such as cell size and apoplastic barriers. (Summary by Magdalena Julkowska) bioRxiv 10.1101/645036
Uptake of water by plants depends on root conductivity, which in turn is determined by hydraulic properties of individual cells and cell anatomy. Quantification of radial root anatomy is a time-consuming process, limiting our understanding of root anatomical features contributing to water uptake. Heymans et al. developed a computational tool Generator of Root ANAtomy in R (GRANAR), which allows for accelerated digitalization of radial root anatomy. The authors successfully re-analyzed maize root anatomies, and by combining GRANAR with MECHA, the hydraulic properties of individual roots could be estimated. GRANAR also allowed predicting conductivity of theoretical root anatomies, revealing the importance of individual root anatomy features, such as cell size and apoplastic barriers. (Summary by Magdalena Julkowska) bioRxiv 10.1101/645036
A growth-based framework for leaf shape development and diversity
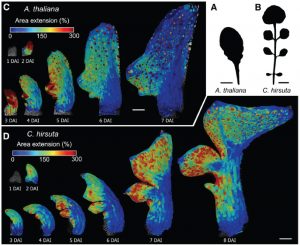 The leaf shape is one of the features defining the diversity in the plant kingdom. However, it is still not understood how action of individual genes is linked to this morphological diversity. Kierzkowski et al. developed an imaging protocol to study the leaf primodium development to understand the cellular growth patterning in two related species showing contrasting leaf morphology – A.thaliana and C.hirsuta. By combining time-lapse imaging, modeling and molecular biology, Kierzkowki et al. identified that the complex leaf shape of C. hirsuta is a result of two processes. The slow and extended growth throughout the primodium and delayed differentiation is regulated by the KNOX gene SHOOTMERISTEMLESS, while local growth inhibition, which emphasizes the marginal serrations, is orchestrated by the REDUCED COMPLEXITY (RCO) gene, which is absent in the A. thaliana genome. (Summary by Magdalena Julkowska) Cell 10.1016/j.cell.2019.05.011
The leaf shape is one of the features defining the diversity in the plant kingdom. However, it is still not understood how action of individual genes is linked to this morphological diversity. Kierzkowski et al. developed an imaging protocol to study the leaf primodium development to understand the cellular growth patterning in two related species showing contrasting leaf morphology – A.thaliana and C.hirsuta. By combining time-lapse imaging, modeling and molecular biology, Kierzkowki et al. identified that the complex leaf shape of C. hirsuta is a result of two processes. The slow and extended growth throughout the primodium and delayed differentiation is regulated by the KNOX gene SHOOTMERISTEMLESS, while local growth inhibition, which emphasizes the marginal serrations, is orchestrated by the REDUCED COMPLEXITY (RCO) gene, which is absent in the A. thaliana genome. (Summary by Magdalena Julkowska) Cell 10.1016/j.cell.2019.05.011
A diversity of traits contributes to salinity tolerance of wild Galapagos tomatoes seedlings
 Domestication has been accompanied by a decrease in genetic diversity, so efforts to improve stress tolerance can be aided by exploring the crop’s wild relatives. Here, Pailles et al. examined salt tolerance in Galapagos tomatoes (Solanum cheesmaniae and Solanum galapagense), which grow “constantly splashed with seawater”. After growing the plants in 200 mM NaCl, the authors quantify eleven traits associated with salinity tolerance in several accessions of these two species, as compared to cultivated Solanum lycopersicum. The data show that the Galapagos tomatoes are more salt tolerant, and that this is associated with traits including potassium levels in leaves, although there is considerable natural variation in the mechanisms of salinity tolerance. One outcome of this work is that it provides a foundation for genetic analysis of these isolates. More broadly, the authors have made freely available all of the data and the tools for analyzing it, making this an excellent training resource. (Summary by Mary Williams) bioRxiv 10.1101/642876
Domestication has been accompanied by a decrease in genetic diversity, so efforts to improve stress tolerance can be aided by exploring the crop’s wild relatives. Here, Pailles et al. examined salt tolerance in Galapagos tomatoes (Solanum cheesmaniae and Solanum galapagense), which grow “constantly splashed with seawater”. After growing the plants in 200 mM NaCl, the authors quantify eleven traits associated with salinity tolerance in several accessions of these two species, as compared to cultivated Solanum lycopersicum. The data show that the Galapagos tomatoes are more salt tolerant, and that this is associated with traits including potassium levels in leaves, although there is considerable natural variation in the mechanisms of salinity tolerance. One outcome of this work is that it provides a foundation for genetic analysis of these isolates. More broadly, the authors have made freely available all of the data and the tools for analyzing it, making this an excellent training resource. (Summary by Mary Williams) bioRxiv 10.1101/642876
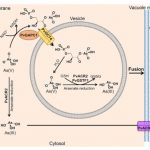
A simple arsenic detoxification strategy in the fern Pteris vittata ($)
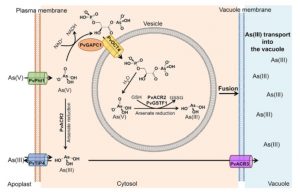 Arsenic contamination is a growing human health threat. The fern Pteris vittata demonstrates a remarkable capacity to accumulate and sequester high levels of the toxic heavy metal arsenic from contaminated environments. Cai et al. used an ‘omics’-guided approach to identify and characterize the molecular mechanisms that afford arsenic tolerance in Pteris. De novo transcriptome analysis of P. vittata gametophytes (haploid life stage) grown with or without arsenate revealed a limited set of arsenate-induced genes that either lacked functional annotation or had predicted roles in transport, phosphate metabolism, or stress responses. Using RNAi-based gene silencing, the authors demonstrated critical roles for three genes in establishing arsenate tolerance in P. vittata; PvGAPC1 (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), PvOCT4 (ORGANIC CATION TRANSPORTER4), and PvGSTF1 (GLUTATHIONE S-TRANSFERASE). Further genetic and biochemical analyses led the authors to propose a likely model for arsenate detoxification in Pteris, which begins with the chemical conversion of arsenate to 1-arseno-3-phosphoglycerate through PvGAPC1, the subsequent transportation of this intermediate into specialized arsenate-metabolizing vesicles through the PvOCT4 transporter, followed by reconversion back to arsenate and chemical reduction to form arsenite that is ultimately transported into the vacuole. This mechanism bears striking resemblance to an arsenic tolerance mechanism described in the Gram-negative bacterium Pseudomonas aeruginosa, which similarly detoxifies arsenic through the 1-arseno-3-phosphoglycerate intermediate. Collectively, the data elegantly illustrate an instance of convergent evolution for the detoxification of arsenic that was independently acquired in ferns and bacteria. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2019.04.029
Arsenic contamination is a growing human health threat. The fern Pteris vittata demonstrates a remarkable capacity to accumulate and sequester high levels of the toxic heavy metal arsenic from contaminated environments. Cai et al. used an ‘omics’-guided approach to identify and characterize the molecular mechanisms that afford arsenic tolerance in Pteris. De novo transcriptome analysis of P. vittata gametophytes (haploid life stage) grown with or without arsenate revealed a limited set of arsenate-induced genes that either lacked functional annotation or had predicted roles in transport, phosphate metabolism, or stress responses. Using RNAi-based gene silencing, the authors demonstrated critical roles for three genes in establishing arsenate tolerance in P. vittata; PvGAPC1 (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), PvOCT4 (ORGANIC CATION TRANSPORTER4), and PvGSTF1 (GLUTATHIONE S-TRANSFERASE). Further genetic and biochemical analyses led the authors to propose a likely model for arsenate detoxification in Pteris, which begins with the chemical conversion of arsenate to 1-arseno-3-phosphoglycerate through PvGAPC1, the subsequent transportation of this intermediate into specialized arsenate-metabolizing vesicles through the PvOCT4 transporter, followed by reconversion back to arsenate and chemical reduction to form arsenite that is ultimately transported into the vacuole. This mechanism bears striking resemblance to an arsenic tolerance mechanism described in the Gram-negative bacterium Pseudomonas aeruginosa, which similarly detoxifies arsenic through the 1-arseno-3-phosphoglycerate intermediate. Collectively, the data elegantly illustrate an instance of convergent evolution for the detoxification of arsenic that was independently acquired in ferns and bacteria. (Summary by Phil Carella) Curr. Biol. 10.1016/j.cub.2019.04.029
Application of TurboID-mediated proximity labeling for mapping a GSK3 Kinase signaling network in Arabidopsis
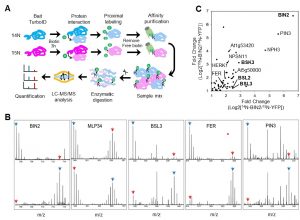 In this preprint Kim et al., have provided an optimized method for determining protein-protein interaction in plants using TurboID-mediated labelling. TurboID is an engineered promiscuous biotin ligase that marks proteins it comes in contact with. In this approach the protein of interest is tagged with a fluorescent marker by TurboID fused to a protein of interest, driven by a constitutive promoter. This allows all the proteins that comes close to the protein of interest to be biotinylated by TurboID more efficiently so that they can can be detected via pull down assays and mass spectrometry. Identification of interacting partners of a protein through this approach will aid in deciphering the molecular signaling network. To evaluate the efficiency of this assay in Arabidopsis the authors have identified the interaction partners of brassinosteroid signaling component, BIN2 (GSK3 like kinase) which will uncover the signaling cascade of this phytohormone. (Summary by Suresh Damodaran) bioRxiv 10.1101/636324
In this preprint Kim et al., have provided an optimized method for determining protein-protein interaction in plants using TurboID-mediated labelling. TurboID is an engineered promiscuous biotin ligase that marks proteins it comes in contact with. In this approach the protein of interest is tagged with a fluorescent marker by TurboID fused to a protein of interest, driven by a constitutive promoter. This allows all the proteins that comes close to the protein of interest to be biotinylated by TurboID more efficiently so that they can can be detected via pull down assays and mass spectrometry. Identification of interacting partners of a protein through this approach will aid in deciphering the molecular signaling network. To evaluate the efficiency of this assay in Arabidopsis the authors have identified the interaction partners of brassinosteroid signaling component, BIN2 (GSK3 like kinase) which will uncover the signaling cascade of this phytohormone. (Summary by Suresh Damodaran) bioRxiv 10.1101/636324
High resolution mapping of RphMBR1012 conferring resistance to Puccinia hordei in barley
 Barley leaf rust (Puccinia hordei) is a biotrophic fungus, in which mutations cause newly virulent races (pathothypes) with severe effects on cultivars. Marker-assisted selection for Rph (Resistance to P. hordei) genes has been studied to provide sustainable control of this disease. Fazlikhani et al. developed molecular markers for the construction of a high resolution map for the RphMBR1012 locus on recombinant inbred lines (RILs), derived from the cross of MBR1012 (resistant) x Scarlett (susceptible) lines. Fifty-six markers were identified/developed using New Illumina SNP genotyping analysis and Genotyping-by-Sequencing (GBS). Marker saturation of the pathogen resistance associated locus and its physical mapping were performed thanks to the barley reference Zipper genome sequence. The authors increased the genetic resolution by the conversion into KASP markers (Kompetitive Allele Specific PCR), which narrowed down the RphMBR1012 target interval to 0.44 Mb between the markers QBS127 and QBS98. In this interval, five candidate R genes were identified and analyzed by allele specific re-sequencing. The markers GBS546 and GBS626 were catalogued as the best to RphMBR1012 detection in barley. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00640.
Barley leaf rust (Puccinia hordei) is a biotrophic fungus, in which mutations cause newly virulent races (pathothypes) with severe effects on cultivars. Marker-assisted selection for Rph (Resistance to P. hordei) genes has been studied to provide sustainable control of this disease. Fazlikhani et al. developed molecular markers for the construction of a high resolution map for the RphMBR1012 locus on recombinant inbred lines (RILs), derived from the cross of MBR1012 (resistant) x Scarlett (susceptible) lines. Fifty-six markers were identified/developed using New Illumina SNP genotyping analysis and Genotyping-by-Sequencing (GBS). Marker saturation of the pathogen resistance associated locus and its physical mapping were performed thanks to the barley reference Zipper genome sequence. The authors increased the genetic resolution by the conversion into KASP markers (Kompetitive Allele Specific PCR), which narrowed down the RphMBR1012 target interval to 0.44 Mb between the markers QBS127 and QBS98. In this interval, five candidate R genes were identified and analyzed by allele specific re-sequencing. The markers GBS546 and GBS626 were catalogued as the best to RphMBR1012 detection in barley. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00640.