Plant Science Research Weekly: May 31, 2024
Review: Strategies to improve photosynthesis
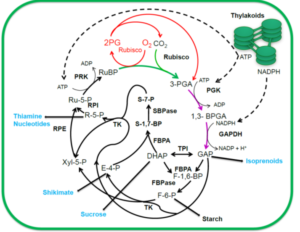 Photosynthesis is the process by which plants assimilate carbon by using light energy. However, with the solar energy conversion efficiency of many crop plants less than 1%, it is inefficient. Therefore, there is interest in manipulating photosynthesis for increased efficiency. Here, Croce et al. identifying nine strategies that have, or could be, implemented to achieve this. At the light absorption level, one strategy is to alter the light that plants absorb to include far-red light, by using cyanobacterial chlorophylls. On the biochemistry side, increasing the expression or activity of sedoheptulose 1,7-bisphosphatase and fructose 1,6-bisphosphate aldolase, which are enzymes of the Calvin-Benson-Bassham cycle, can increase photosynthesis. The authors also suggest some high-risk, high-reward strategies for increasing the activity of RuBisCo, a major rate limiting enzyme in the Calvin-Benson-Bassham cycle. These include engineering carbon concentrating mechanisms from green algae into plants, or replacing native RuBisCo with a higher efficiency version found in red algae. Finally, they consider leaf physiology, such as how increasing stomatal density can increase photosynthesis, although this can also lead to increased water loss. Overall, there are a plethora of strategies which could be used to improve photosynthesis, and these need to be integrated together for the maximum efficiency. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae132
Photosynthesis is the process by which plants assimilate carbon by using light energy. However, with the solar energy conversion efficiency of many crop plants less than 1%, it is inefficient. Therefore, there is interest in manipulating photosynthesis for increased efficiency. Here, Croce et al. identifying nine strategies that have, or could be, implemented to achieve this. At the light absorption level, one strategy is to alter the light that plants absorb to include far-red light, by using cyanobacterial chlorophylls. On the biochemistry side, increasing the expression or activity of sedoheptulose 1,7-bisphosphatase and fructose 1,6-bisphosphate aldolase, which are enzymes of the Calvin-Benson-Bassham cycle, can increase photosynthesis. The authors also suggest some high-risk, high-reward strategies for increasing the activity of RuBisCo, a major rate limiting enzyme in the Calvin-Benson-Bassham cycle. These include engineering carbon concentrating mechanisms from green algae into plants, or replacing native RuBisCo with a higher efficiency version found in red algae. Finally, they consider leaf physiology, such as how increasing stomatal density can increase photosynthesis, although this can also lead to increased water loss. Overall, there are a plethora of strategies which could be used to improve photosynthesis, and these need to be integrated together for the maximum efficiency. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae132
Review: Optimizing nutrient transporters to enhance disease resistance in rice
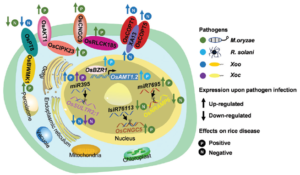 Plants rely on an array of mineral nutrients for their growth, development, and reproductive processes. The molecular mechanisms governing the uptake, translocation, storage, and utilization of these essential minerals are orchestrated by specific nutrient transporters and their associated regulatory elements, including microRNAs and the ubiquitylation system. Intriguingly, mounting evidence indicates that beyond the traditional roles in plant physiology, mineral nutrients also play pivotal roles in bolstering plant defense mechanisms against pathogens. On one front, the modulation of plant nutrient status, achieved through adjustments in external nutrient availability or the manipulation of nutrient transporter expression, can significantly impact the output of the plant’s immune response. Conversely, the expression levels of nutrient transporters themselves can be influenced by pathogen invasion. This intricate bi-directional interplay underscores the close nexus between nutrient status and the efficacy of plant immunity. Hui and colleagues offer a comprehensive review of existing literature, elucidating the connections between nutrient transporters and disease resistance in rice. Their work not only underscores the pivotal role of nutrient transporters in fortifying plant defenses but also presents promising avenues for enhancing disease resistance in rice through genetic engineering strategies targeting these transporters. (Summary by Ching Chan @ntnuchanlab) J. Exp. Bot. 10.1093/jxb/erae087
Plants rely on an array of mineral nutrients for their growth, development, and reproductive processes. The molecular mechanisms governing the uptake, translocation, storage, and utilization of these essential minerals are orchestrated by specific nutrient transporters and their associated regulatory elements, including microRNAs and the ubiquitylation system. Intriguingly, mounting evidence indicates that beyond the traditional roles in plant physiology, mineral nutrients also play pivotal roles in bolstering plant defense mechanisms against pathogens. On one front, the modulation of plant nutrient status, achieved through adjustments in external nutrient availability or the manipulation of nutrient transporter expression, can significantly impact the output of the plant’s immune response. Conversely, the expression levels of nutrient transporters themselves can be influenced by pathogen invasion. This intricate bi-directional interplay underscores the close nexus between nutrient status and the efficacy of plant immunity. Hui and colleagues offer a comprehensive review of existing literature, elucidating the connections between nutrient transporters and disease resistance in rice. Their work not only underscores the pivotal role of nutrient transporters in fortifying plant defenses but also presents promising avenues for enhancing disease resistance in rice through genetic engineering strategies targeting these transporters. (Summary by Ching Chan @ntnuchanlab) J. Exp. Bot. 10.1093/jxb/erae087
Review: Proteolysis in plant immunity
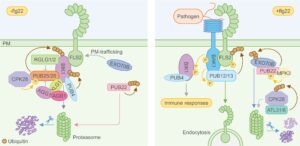 Plant immunity is beautifully sophisticated with a ever-growing cast of characters. This review by Liu et al. focuses on the contributions of proteolysis (protein degradation), which both activates and suppresses immune responses. The authors structure the review around four key functions of proteolysis: direct degradation of pathogen proteins, activation of host immune proteins, regulation of immune component stability, and release of immunogenic peptides. They next turn to systems that are regulated through the ubiquitin-proteasome system, surveying its roles in PRR (pattern-recognition receptor) and NLR (nucleotide binding leucine-rich repeat receptor) immunity, as well as in salicylic acid signaling. They conclude by summarizing efforts to engineer proteolysis to enhance immunity and by highlighting open questions. It’s a must-read resource for those working in the field and those who strive to keep up to date with this fast-moving topic. (Summary by Mary Williams @PlantTeaching) Plant Cell https://doi.org/10.1093/plcell/koae142
Plant immunity is beautifully sophisticated with a ever-growing cast of characters. This review by Liu et al. focuses on the contributions of proteolysis (protein degradation), which both activates and suppresses immune responses. The authors structure the review around four key functions of proteolysis: direct degradation of pathogen proteins, activation of host immune proteins, regulation of immune component stability, and release of immunogenic peptides. They next turn to systems that are regulated through the ubiquitin-proteasome system, surveying its roles in PRR (pattern-recognition receptor) and NLR (nucleotide binding leucine-rich repeat receptor) immunity, as well as in salicylic acid signaling. They conclude by summarizing efforts to engineer proteolysis to enhance immunity and by highlighting open questions. It’s a must-read resource for those working in the field and those who strive to keep up to date with this fast-moving topic. (Summary by Mary Williams @PlantTeaching) Plant Cell https://doi.org/10.1093/plcell/koae142
Translocation of a chloride channel from the Golgi to the plasma membrane helps plants adapt to salt stress
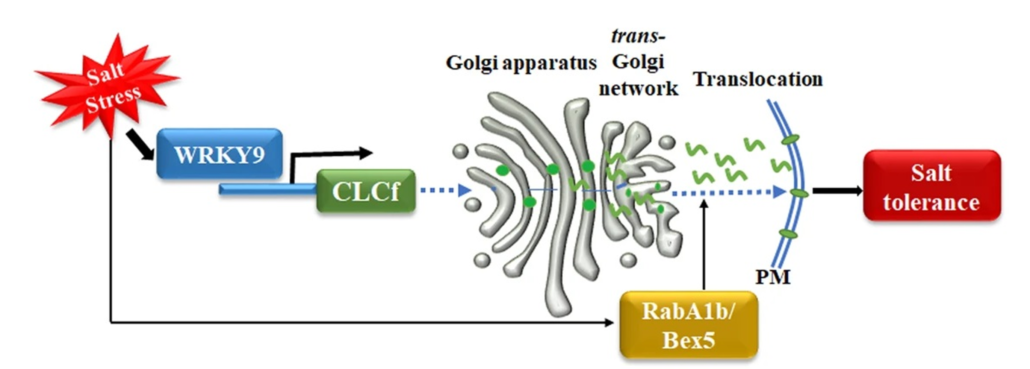 Plants adapt to salt stress by maintaining ion balance using ion transporters. While much is known about cation transporters, the role of anion transporters is less clear. Rajappa et al. focus on the chloride channel gene AtCLCf in Arabidopsis thaliana, controlled by the transcription factor WRKY9 under salt stress. Plants lacking AtCLCf are more sensitive to salt, while those overexpressing it are more resistant. Salt stress causes the AtCLCf protein to move to the plasma membrane, and inhibiting this movement increases salt sensitivity. Experiments confirm that AtCLCf functions as a Cl−/H+ antiporter on the Golgi apparatus. This discovery reveals that AtCLCf helps plants cope with salt stress by removing chloride ions from the roots, thus enhancing salt tolerance. (Summary by Yueh Cho @YuehCho1984) 10.1038/s41467-024-48234-z
Plants adapt to salt stress by maintaining ion balance using ion transporters. While much is known about cation transporters, the role of anion transporters is less clear. Rajappa et al. focus on the chloride channel gene AtCLCf in Arabidopsis thaliana, controlled by the transcription factor WRKY9 under salt stress. Plants lacking AtCLCf are more sensitive to salt, while those overexpressing it are more resistant. Salt stress causes the AtCLCf protein to move to the plasma membrane, and inhibiting this movement increases salt sensitivity. Experiments confirm that AtCLCf functions as a Cl−/H+ antiporter on the Golgi apparatus. This discovery reveals that AtCLCf helps plants cope with salt stress by removing chloride ions from the roots, thus enhancing salt tolerance. (Summary by Yueh Cho @YuehCho1984) 10.1038/s41467-024-48234-z
Alone or together: BRI1 signaling controls root growth in a cell-autonomous manner
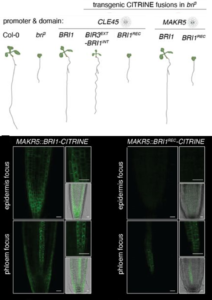 Root growth is controlled by a dense network of hormone signals working in shared and distinct root zones and cell types. Brassinosteroids (BR) control root growth through activated signaling in vascular and epidermal tissues, but it is unclear how BR signals in each tissue contribute and whether there are cell non-autonomous effects. Using transgenic lines deficient in BR perception (bri3) but complemented with a functional BRI1 under the control of cell-type-specific promoters, Blanco- Touriñán et al. revealed that BR perception in phloem cells is largely sufficient to rescue root growth. In these plants, fluorescently labeled BRI1 revealed that BRI1 is not only present in phloem cells but is also ectopically expressed in the epidermis and other cell types, though at low levels. Employing a recoded but functional version of BRI1 (BRI1REC), the authors demonstrated that specific intrinsic regulatory sequences within the BRI1 gene body, specifically the coding region, are crucial for this ubiquitous low-level expression and are required for a proper rescue of the bri3 mutant phenotype. Such an effect of a gene primary structure controlling its expression patterns was relatively unknown and is an interesting topic for further studies. To conclude, the authors elegantly demonstrated a mechanism of BR signaling through its receptors in a cell-autonomous manner, aided by the intrinsic regulatory expression mechanisms of the genes. (Summary by Thomas Depaepe @thdpaepe) bioRxiv 10.1101/2024.05.13.593848.
Root growth is controlled by a dense network of hormone signals working in shared and distinct root zones and cell types. Brassinosteroids (BR) control root growth through activated signaling in vascular and epidermal tissues, but it is unclear how BR signals in each tissue contribute and whether there are cell non-autonomous effects. Using transgenic lines deficient in BR perception (bri3) but complemented with a functional BRI1 under the control of cell-type-specific promoters, Blanco- Touriñán et al. revealed that BR perception in phloem cells is largely sufficient to rescue root growth. In these plants, fluorescently labeled BRI1 revealed that BRI1 is not only present in phloem cells but is also ectopically expressed in the epidermis and other cell types, though at low levels. Employing a recoded but functional version of BRI1 (BRI1REC), the authors demonstrated that specific intrinsic regulatory sequences within the BRI1 gene body, specifically the coding region, are crucial for this ubiquitous low-level expression and are required for a proper rescue of the bri3 mutant phenotype. Such an effect of a gene primary structure controlling its expression patterns was relatively unknown and is an interesting topic for further studies. To conclude, the authors elegantly demonstrated a mechanism of BR signaling through its receptors in a cell-autonomous manner, aided by the intrinsic regulatory expression mechanisms of the genes. (Summary by Thomas Depaepe @thdpaepe) bioRxiv 10.1101/2024.05.13.593848.
COP1 regulates salt tolerance via GIGANTEA degradation in roots
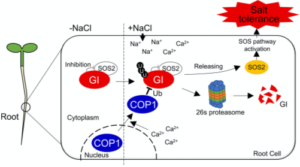 Global climate change affects weather patterns, and excess soil salinity harms plant growth. Previous studies have shown interaction between the circadian clock and salt responses. In normal conditions, the core circadian clock oscillator GIGANTEA (GI) sequesters the SOS2 kinase, part of the salt-overly sensitive (SOS) pathway. Salt stress triggers calcium signals that cause GI to break down in the cytoplasm via the 26S proteasome, activating the SOS pathway. A new study by Ji and Khakurel et al. reveals that these calcium signals lead to the degradation of GI exclusively in roots through the action of the E3 ubiquitin ligase COP1 protein. Under salt stress, COP1 moves to the cytoplasm where it targets GI for proteasomal destruction. COP1 has both nuclear- and cytoplasmic-localization signals, so selectively mutating these revealed that COP1 restricted to the nucleus leads to enhanced salt sensitivity, and COP1 restricted to the cytoplasm enhances salt tolerance. Furthermore, plants with mutations in both GI and COP1 show increased salt tolerance, highlighting the key role of GI. This research sheds light on the roles of COP1 and GI in salt tolerance, offering new avenues for creating salt-tolerant crops. (Summary by Yueh Cho @YuehCho1984) Plant Cell Environ 10.1111/pce.14946.
Global climate change affects weather patterns, and excess soil salinity harms plant growth. Previous studies have shown interaction between the circadian clock and salt responses. In normal conditions, the core circadian clock oscillator GIGANTEA (GI) sequesters the SOS2 kinase, part of the salt-overly sensitive (SOS) pathway. Salt stress triggers calcium signals that cause GI to break down in the cytoplasm via the 26S proteasome, activating the SOS pathway. A new study by Ji and Khakurel et al. reveals that these calcium signals lead to the degradation of GI exclusively in roots through the action of the E3 ubiquitin ligase COP1 protein. Under salt stress, COP1 moves to the cytoplasm where it targets GI for proteasomal destruction. COP1 has both nuclear- and cytoplasmic-localization signals, so selectively mutating these revealed that COP1 restricted to the nucleus leads to enhanced salt sensitivity, and COP1 restricted to the cytoplasm enhances salt tolerance. Furthermore, plants with mutations in both GI and COP1 show increased salt tolerance, highlighting the key role of GI. This research sheds light on the roles of COP1 and GI in salt tolerance, offering new avenues for creating salt-tolerant crops. (Summary by Yueh Cho @YuehCho1984) Plant Cell Environ 10.1111/pce.14946.
Glutamine induces lateral root initiation, stress responses, and disease resistance in Arabidopsis
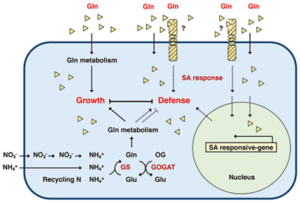 Nitrogen (N) is vital for plant growth, with plants typically absorbing inorganic N compounds like nitrate (NO3−) and ammonium (NH4+) from the soil. Yet plants can also utilize organic N sources, including amino acids like glutamine (Gln). Gln, as the first organic N compound assimilated within plant cells, goes beyond its metabolic function, influencing various aspects of plant biology. It regulates cytokinin biosynthesis, maintains salicylic acid (SA)-associated redox equilibrium, and plays roles in stress responses and pathogen defense, making it a key signaling molecule in plants. In a study conducted by Liao and colleagues, supplementing Gln as the sole nitrogen source not only stimulated plant growth and lateral root initiation, but also upregulated stress-responsive genes. Intriguingly, exogenous Gln primed the expression of defense genes, which in turn, reduced bacterial proliferation upon Pst DC3000 infection in Arabidopsis. Notably, this protective effect was abolished in the sid2 mutant and partially attenuated in the npr1 mutant, highlighting the reliance of Gln-mediated immune responses on the salicylic acid signaling pathway. This study sheds light on the dual nature of Gln, serving as both a metabolic derivative and a critical component in immune signaling pathways within Arabidopsis, which unveils the intricate interplay between nitrogen metabolism and plant defense mechanisms. (Summary by Ching Chan @ntnuchanlab) Plant Physiol. 10.1093/plphys/kiae144
Nitrogen (N) is vital for plant growth, with plants typically absorbing inorganic N compounds like nitrate (NO3−) and ammonium (NH4+) from the soil. Yet plants can also utilize organic N sources, including amino acids like glutamine (Gln). Gln, as the first organic N compound assimilated within plant cells, goes beyond its metabolic function, influencing various aspects of plant biology. It regulates cytokinin biosynthesis, maintains salicylic acid (SA)-associated redox equilibrium, and plays roles in stress responses and pathogen defense, making it a key signaling molecule in plants. In a study conducted by Liao and colleagues, supplementing Gln as the sole nitrogen source not only stimulated plant growth and lateral root initiation, but also upregulated stress-responsive genes. Intriguingly, exogenous Gln primed the expression of defense genes, which in turn, reduced bacterial proliferation upon Pst DC3000 infection in Arabidopsis. Notably, this protective effect was abolished in the sid2 mutant and partially attenuated in the npr1 mutant, highlighting the reliance of Gln-mediated immune responses on the salicylic acid signaling pathway. This study sheds light on the dual nature of Gln, serving as both a metabolic derivative and a critical component in immune signaling pathways within Arabidopsis, which unveils the intricate interplay between nitrogen metabolism and plant defense mechanisms. (Summary by Ching Chan @ntnuchanlab) Plant Physiol. 10.1093/plphys/kiae144
REF1 peptide is a wound signal that promotes plant regeneration
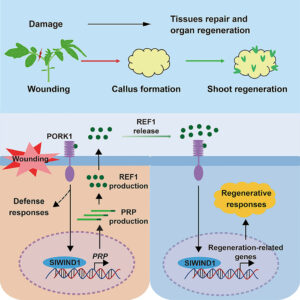 Plants have a remarkable ability to reprogram and regenerate as needed following wounding. This trait is invaluable not only to gardeners but also to regenerate plants following genome editing. Here, Wang, Zhai, Wu, and Deng have identified key steps in tissue regeneration. Through characterization of a tomato mutant with deficiencies in wound-induced regeneration, they identified a gene encoding a precursor protein that is cleaved to produce a peptide corresponding to Arabidopsis Pep6, which they subsequently named REF1 peptide due to its role in regeneration. Application of REF1 to plant tissues induces the wound response, as assessed by expression of the proteinase inhibitor marker PI-II. Plants overexpressing the precursor of REF1 also show enhanced callus and shoot regeneration in tissue culture. By homology to Arabidopsis, the authors identified a LRR-receptor kinase called PORK1 (PEPR1/2-ORTHOLOG-RECEPTOR-LIKE KINASE1) that is necessary for responses to REF1. Again by homology to Arabidopsis, they showed that the REF1-PORK1 signaling module works through the transcription factor WIND1. Finally, they showed that application of REF1 can promote tissue regeneration of soybean, wheat, and maize. Because tissue regeneration can be a significant obstacle to employing gene editing technologies to many crops, these findings will have a big impact. (Summary by Mary Williams @PlantTeaching) Cell https://doi.org/10.1016/j.cell.2024.04.040
Plants have a remarkable ability to reprogram and regenerate as needed following wounding. This trait is invaluable not only to gardeners but also to regenerate plants following genome editing. Here, Wang, Zhai, Wu, and Deng have identified key steps in tissue regeneration. Through characterization of a tomato mutant with deficiencies in wound-induced regeneration, they identified a gene encoding a precursor protein that is cleaved to produce a peptide corresponding to Arabidopsis Pep6, which they subsequently named REF1 peptide due to its role in regeneration. Application of REF1 to plant tissues induces the wound response, as assessed by expression of the proteinase inhibitor marker PI-II. Plants overexpressing the precursor of REF1 also show enhanced callus and shoot regeneration in tissue culture. By homology to Arabidopsis, the authors identified a LRR-receptor kinase called PORK1 (PEPR1/2-ORTHOLOG-RECEPTOR-LIKE KINASE1) that is necessary for responses to REF1. Again by homology to Arabidopsis, they showed that the REF1-PORK1 signaling module works through the transcription factor WIND1. Finally, they showed that application of REF1 can promote tissue regeneration of soybean, wheat, and maize. Because tissue regeneration can be a significant obstacle to employing gene editing technologies to many crops, these findings will have a big impact. (Summary by Mary Williams @PlantTeaching) Cell https://doi.org/10.1016/j.cell.2024.04.040
Coordinated wound responses in a regenerative animal-algal holobiont
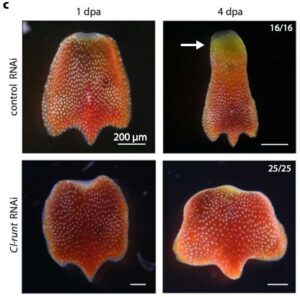 Many animals such as corals and sea slugs host photosynthetic symbionts. Several studies have investigated how the host and symbiont coordinate their activities. This new study by Sarfati et al. explores how symbiotic green algae and their animal host, the flattened-worm Convolutriloba longifissura, respond to injury. When the animal is injured, the algae’s photosynthetic efficiency drops, and the animal and algae show two waves of gene activity. Initially, the algae increase the activity of genes related to photosynthesis, although photosynthesis is not needed for regeneration. An important animal gene, runt, is activated after injury and is necessary for the animal’s regeneration. Decreasing runt expression alters the transcriptional responses in both the animal and algae and further decreases the photosynthesis by the algae after injury. This research shows that the host and symbionts are integrated at a molecular level and function as an integrated biological unit during regeneration. (Summary by Yueh Cho @YuehCho1984) Nature Comms. 10.1038/s41467-024-48366-2.
Many animals such as corals and sea slugs host photosynthetic symbionts. Several studies have investigated how the host and symbiont coordinate their activities. This new study by Sarfati et al. explores how symbiotic green algae and their animal host, the flattened-worm Convolutriloba longifissura, respond to injury. When the animal is injured, the algae’s photosynthetic efficiency drops, and the animal and algae show two waves of gene activity. Initially, the algae increase the activity of genes related to photosynthesis, although photosynthesis is not needed for regeneration. An important animal gene, runt, is activated after injury and is necessary for the animal’s regeneration. Decreasing runt expression alters the transcriptional responses in both the animal and algae and further decreases the photosynthesis by the algae after injury. This research shows that the host and symbionts are integrated at a molecular level and function as an integrated biological unit during regeneration. (Summary by Yueh Cho @YuehCho1984) Nature Comms. 10.1038/s41467-024-48366-2.
Induction of disease suppressive microbiome by non-pathogenic bacterial derivatives
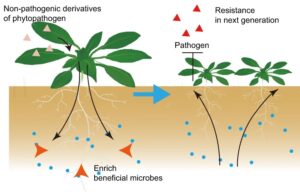 Plants recruit beneficial microbiomes in the rhizosphere in response to aboveground pathogen infections, a mechanism known as “cry for help.” Zhang and Liu et al. investigated this effect to test whether pathogenicity is necessary for assembling a disease-suppressive rhizomicrobiome. By using nonpathogenic derivatives of Pseudomonas syringae (Pst) DC3000 to elicit immune responses in Arabidopsis, the authors identified two derivatives that induce disease suppression similar to the wild-type Pst DC3000. Importantly, these derivatives only triggered pattern-triggered immunity (PTI) and reactive oxygen species (ROS) production without activating effector-triggered immunity (ETI). Analyses of the rhizosphere microbiome infiltrated with these non-pathogenic derivatives identified Devosia as a key enriched taxon. Interestingly, the application of cultured consortia of two Devosia strains reduced the colonization of pathogenic Pst DC3000 and enhanced plant fresh weight. Furthermore, the researchers identified a decrease in the exudation of myristic acid as the key metabolite favoring the enrichment of these specific strains in the rhizosphere. Finally, the authors discovered that secreted metabolites from these non-pathogenic derivatives are sufficient to induce a “cry for help”, which could be blocked by ETI. This study suggests the possibility of identifying non-pathogenic elicitors to repress plant diseases without growth penalties. (Summary by Arijit Mukherjee @ArijitM61745830) Nature Comms. https://doi.org/10.1038/s41467-024-46254-3
Plants recruit beneficial microbiomes in the rhizosphere in response to aboveground pathogen infections, a mechanism known as “cry for help.” Zhang and Liu et al. investigated this effect to test whether pathogenicity is necessary for assembling a disease-suppressive rhizomicrobiome. By using nonpathogenic derivatives of Pseudomonas syringae (Pst) DC3000 to elicit immune responses in Arabidopsis, the authors identified two derivatives that induce disease suppression similar to the wild-type Pst DC3000. Importantly, these derivatives only triggered pattern-triggered immunity (PTI) and reactive oxygen species (ROS) production without activating effector-triggered immunity (ETI). Analyses of the rhizosphere microbiome infiltrated with these non-pathogenic derivatives identified Devosia as a key enriched taxon. Interestingly, the application of cultured consortia of two Devosia strains reduced the colonization of pathogenic Pst DC3000 and enhanced plant fresh weight. Furthermore, the researchers identified a decrease in the exudation of myristic acid as the key metabolite favoring the enrichment of these specific strains in the rhizosphere. Finally, the authors discovered that secreted metabolites from these non-pathogenic derivatives are sufficient to induce a “cry for help”, which could be blocked by ETI. This study suggests the possibility of identifying non-pathogenic elicitors to repress plant diseases without growth penalties. (Summary by Arijit Mukherjee @ArijitM61745830) Nature Comms. https://doi.org/10.1038/s41467-024-46254-3
The Nagoya Protocol and nitrogen-fixing maize: Close encounters between Indigenous Oaxacans and the men from Mars (Inc.)
 Most readers recognize the photograph of the amazing nitrogen-fixing maize, with its red, slimy aerial roots that nurture nitrogen-fixing bacteria. But I suspect few of us have thought about how this landrace came to the attention of the mainstream media, who owns the rights to it, and how it would be exploited and by whom. Those stories are found in this new article by Kloppenburg et al., which uses it as a case study to explore the wrong and right way to recognize and compensate Indigenous and local people for their natural resources and knowledge. The history of plant science is replete with knowledge and plant material taken without credit, recognition, or compensation (see Mabry et al., 2023), a practice sometimes called biopiracy, but many of these stories are from the distant past. What makes this article particularly relevant and important for teaching is that it describes a problematic and a very recent occurrence, one post-awareness of biopiracy and after adoption of the Nagoya Protocol (fully, the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from Their Utilization). Although the nitrogen-fixing maize was accessed and utilized in compliance with the Nagoya Protocol, the authors point out that the process was neither equitable nor fair to those who have cultivated it for generations. The article presents the process through which the agreement was made, highlighting the ambiguities of Nagoya, and offers alternative strategies to allow this intriguing and potentially impactful trait to be studied. I encourage you to read this, and particularly to share this important lesson with students. (Summary by Mary Williams @PlantTeaching) Elemanta https://doi.org/10.1525/elementa.2023.00115
Most readers recognize the photograph of the amazing nitrogen-fixing maize, with its red, slimy aerial roots that nurture nitrogen-fixing bacteria. But I suspect few of us have thought about how this landrace came to the attention of the mainstream media, who owns the rights to it, and how it would be exploited and by whom. Those stories are found in this new article by Kloppenburg et al., which uses it as a case study to explore the wrong and right way to recognize and compensate Indigenous and local people for their natural resources and knowledge. The history of plant science is replete with knowledge and plant material taken without credit, recognition, or compensation (see Mabry et al., 2023), a practice sometimes called biopiracy, but many of these stories are from the distant past. What makes this article particularly relevant and important for teaching is that it describes a problematic and a very recent occurrence, one post-awareness of biopiracy and after adoption of the Nagoya Protocol (fully, the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from Their Utilization). Although the nitrogen-fixing maize was accessed and utilized in compliance with the Nagoya Protocol, the authors point out that the process was neither equitable nor fair to those who have cultivated it for generations. The article presents the process through which the agreement was made, highlighting the ambiguities of Nagoya, and offers alternative strategies to allow this intriguing and potentially impactful trait to be studied. I encourage you to read this, and particularly to share this important lesson with students. (Summary by Mary Williams @PlantTeaching) Elemanta https://doi.org/10.1525/elementa.2023.00115