Plant Science Research Weekly: May 29th
Review: The bHLH network underlying plant shade-avoidance
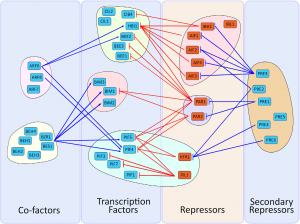 Shade avoidance is a complex phenomenon in which plants avoid shade by altering their developmental program in various ways including early flowering, hypocotyl elongation, and more. Many photoreceptors and transcription factors (TFs) are involved in regulating shade avoidance, including the bHLH (basic helix loop helix) TF family that comprises about 160 members in Arabidopsis. In this review, Buti et al. highlight the contributions of bHLH proteins during shade conditions. They focus on PIF (Phytochrome interacting factors), BEE (Brassinosteroid enhanced expression), BIM (BES1-interacting MIC-1), PAR (Phytochrome-rapidly regulated), and AIF (ATBS1 interacting factor) subfamilies (and they include a helpful table that lists the subfamilies, bHLH name, gene name, and which other proteins each interacts with). Studies have shown that members of the bHLH family either directly bind DNA or indirectly bind DNA by associating with other bHLH family members or other cofactors to regulate downstream gene expression. The authors also describe the role of photoreceptors in controlling the activity of bHLH sub-families during shade conditions. This review will be a great resource for understanding gene regulation during different light conditions. (Summary by Sunita Pathak @psunita980) Physiologia Plantarum 10.1111/ppl.13074
Shade avoidance is a complex phenomenon in which plants avoid shade by altering their developmental program in various ways including early flowering, hypocotyl elongation, and more. Many photoreceptors and transcription factors (TFs) are involved in regulating shade avoidance, including the bHLH (basic helix loop helix) TF family that comprises about 160 members in Arabidopsis. In this review, Buti et al. highlight the contributions of bHLH proteins during shade conditions. They focus on PIF (Phytochrome interacting factors), BEE (Brassinosteroid enhanced expression), BIM (BES1-interacting MIC-1), PAR (Phytochrome-rapidly regulated), and AIF (ATBS1 interacting factor) subfamilies (and they include a helpful table that lists the subfamilies, bHLH name, gene name, and which other proteins each interacts with). Studies have shown that members of the bHLH family either directly bind DNA or indirectly bind DNA by associating with other bHLH family members or other cofactors to regulate downstream gene expression. The authors also describe the role of photoreceptors in controlling the activity of bHLH sub-families during shade conditions. This review will be a great resource for understanding gene regulation during different light conditions. (Summary by Sunita Pathak @psunita980) Physiologia Plantarum 10.1111/ppl.13074
Nutrient dose-responsive transcriptome changes driven by Michaelis–Menten kinetics underlie plant growth rates
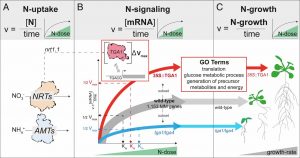 Plants can increase their growth and biomass proportionately to an increase in nutrient dose and, conversely, their growth is limited by limiting nutrients. In this study, Swift et al. explored the molecular underpinnings of the nutrient dose-response phenomenon. The authors first show that nitrogen-dose affects the rate of transcript change genome-wide in Arabidopsis. They showed that this dose-responsive gene expression pattern follows the Michaelis-Menten (MM) kinetics model. However, this relationship is not linear because at higher doses of nitrogen there was a diminishing effect on the rate at which genes were induced or repressed. Since transcription factors largely controls gene expression, the authors reason that transcription factors could be acting as a catalyst driving the rate of transcript change in response to nitrogen-dose. The authors identified three transcription factors; LBD37/38 and TGA1 as candidate transcriptional regulators of the nitrogen-dose response. They looked for cis-regulatory elements overrepresented in the promoter of genes whose transcript change in response to nitrogen-dose follow the MM model and found that the TGA1 binding site was overrepresented. In addition, those genes are also preferentially expressed in the root pericycle and stele where TGA1 itself is expressed. Using knock out mutants and overexpression lines, they showed that TGA1 levels mediate accelerated Arabidopsis growth rates in response to nitrogen dose. (Summary by Toluwase Olukayodo @toluxylic) Proc. Natl. Acad. Sci. USA
Plants can increase their growth and biomass proportionately to an increase in nutrient dose and, conversely, their growth is limited by limiting nutrients. In this study, Swift et al. explored the molecular underpinnings of the nutrient dose-response phenomenon. The authors first show that nitrogen-dose affects the rate of transcript change genome-wide in Arabidopsis. They showed that this dose-responsive gene expression pattern follows the Michaelis-Menten (MM) kinetics model. However, this relationship is not linear because at higher doses of nitrogen there was a diminishing effect on the rate at which genes were induced or repressed. Since transcription factors largely controls gene expression, the authors reason that transcription factors could be acting as a catalyst driving the rate of transcript change in response to nitrogen-dose. The authors identified three transcription factors; LBD37/38 and TGA1 as candidate transcriptional regulators of the nitrogen-dose response. They looked for cis-regulatory elements overrepresented in the promoter of genes whose transcript change in response to nitrogen-dose follow the MM model and found that the TGA1 binding site was overrepresented. In addition, those genes are also preferentially expressed in the root pericycle and stele where TGA1 itself is expressed. Using knock out mutants and overexpression lines, they showed that TGA1 levels mediate accelerated Arabidopsis growth rates in response to nitrogen dose. (Summary by Toluwase Olukayodo @toluxylic) Proc. Natl. Acad. Sci. USA
How do auxin temporal dynamics regulate patterning?
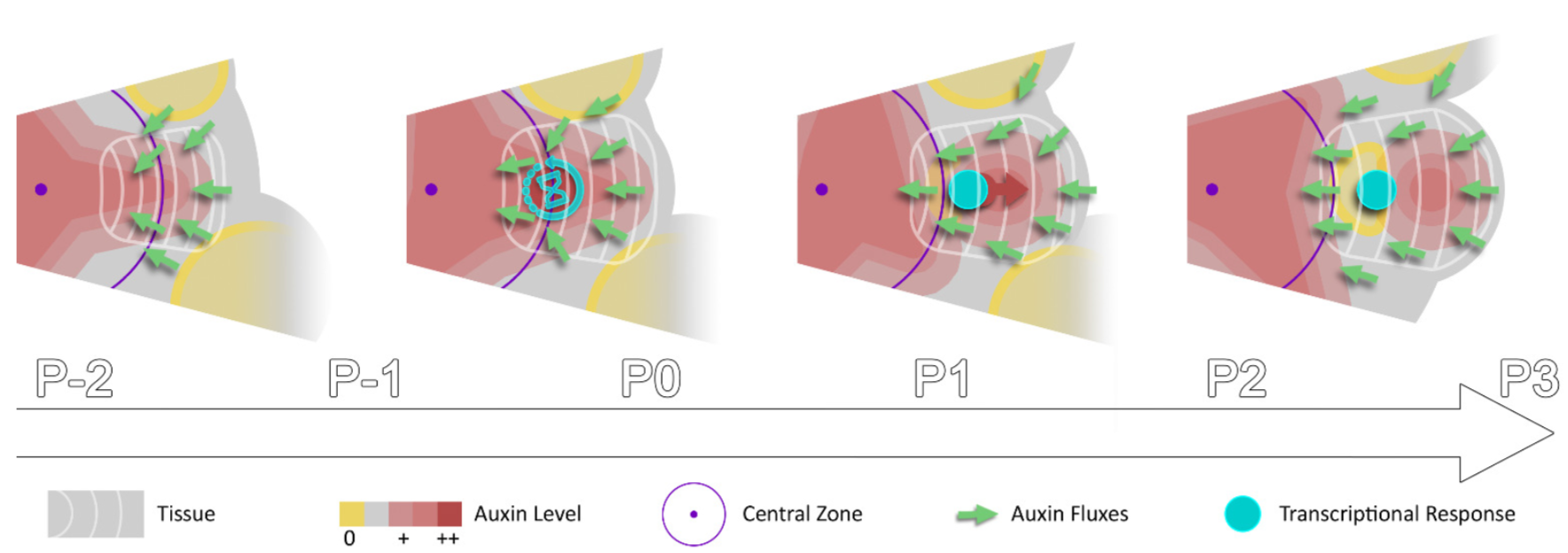 Auxin forms spatial gradients that are implicated in organ morphogenesis. However, it is not known how temporal auxin gradients are integrated with the spatial information. In this paper, Galvan-Ampudia, Cerutti et al., showed using a ratiometric quantitative auxin reporter (quantitative DII-VENUS) that auxin moves faster than cellular growth in the shoot apical meristem (SAM), and that the auxin maximum is not linked to specific cells but travels through the tissue radially in centrifugal manner producing a rhythmic pattern. The authors analyzed orientation of the auxin efflux protein PIN FORMED1 (PIN1) and observed a that the net orientation is towards the center of the SAM raising the question where and when the auxin synthesis happens. Analysis of YUCCA genes encoding auxin biosynthetic enzymes showed that the auxin synthesis happens in the primordia that are more developmentally advanced than the incipient primordia and is channeled to central zone of the SAM by the orientation of the PIN1 protein. Surprisingly, the authors observed that the auxin maximum does not immediately translate into transcriptional responses as there is a delay in the expression of an auxin sensitive transcriptional reporter (DR5) indicating that cells doesn’t become competent to auxin response until 10-14 hrs later (the time between successive organs). Thus, this paper shows that temporal auxin information is integrated in space for organogenesis expanding our understanding of auxin dynamics in time and space. (Summary by Vijaya Batthula @Vijaya_Batthula). eLife 10.7554/eLife.55832.
Auxin forms spatial gradients that are implicated in organ morphogenesis. However, it is not known how temporal auxin gradients are integrated with the spatial information. In this paper, Galvan-Ampudia, Cerutti et al., showed using a ratiometric quantitative auxin reporter (quantitative DII-VENUS) that auxin moves faster than cellular growth in the shoot apical meristem (SAM), and that the auxin maximum is not linked to specific cells but travels through the tissue radially in centrifugal manner producing a rhythmic pattern. The authors analyzed orientation of the auxin efflux protein PIN FORMED1 (PIN1) and observed a that the net orientation is towards the center of the SAM raising the question where and when the auxin synthesis happens. Analysis of YUCCA genes encoding auxin biosynthetic enzymes showed that the auxin synthesis happens in the primordia that are more developmentally advanced than the incipient primordia and is channeled to central zone of the SAM by the orientation of the PIN1 protein. Surprisingly, the authors observed that the auxin maximum does not immediately translate into transcriptional responses as there is a delay in the expression of an auxin sensitive transcriptional reporter (DR5) indicating that cells doesn’t become competent to auxin response until 10-14 hrs later (the time between successive organs). Thus, this paper shows that temporal auxin information is integrated in space for organogenesis expanding our understanding of auxin dynamics in time and space. (Summary by Vijaya Batthula @Vijaya_Batthula). eLife 10.7554/eLife.55832.
Design principles of a minimal auxin response system
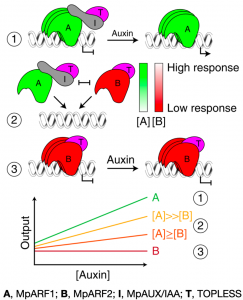 Auxin controls virtually all facets of growth and development. This plant hormone is sensed by TIR1/AFB F-box receptors which promote ubiquitin-mediated degradation of AUX/IAA transcriptional repressors, releasing ARF transcription factors from inhibition. Phylogenetic analyses divide the ARF family into three conserved classes: A/B/C, where A-ARFs act as activators, with some B and C-ARFs acting as repressors. However, despite the apparent simplicity of this signaling module, each member belongs to gene families that are expanded in most land plants (up to 30 ARFs in Arabidopsis) and therefore may have neo- or sub- functionalized across evolution, acquiring different biochemical properties and functions, thus making the deduction of an ancestral auxin response system difficult. A recent study by Kato et al. took advantage of the liverwort Marchantia polymorpha low genetic redundancy (one TIR/AFB receptor, one AUX/IAA and three ARFs, A/B/C) to uncover the biochemical basis of a minimal auxin response system. Using a combination of molecular, structural and bioinformatic approaches, the authors deduced the M. polymorpha auxin response system, where a single A-ARF (MpARF1) switches from Aux/IAA-mediated repression to transcriptional activator in an auxin-dependent manner, whereas an auxin-insensitive B-ARF (MpARF2) antagonizes MpARF1, competing for the same target sites and acting as a repressor by recruiting the TOPLESS (TPL) corepressor. However, differential spatial distribution patterns influence the stoichiometry of these proteins, therefore resulting in the creation of zones with different auxin sensitivities to fine-tune growth and development. The consistency of this model in more complex lineages, such as Physcomitrella or Arabidopsis, suggests that this simple module may be the chassis for more complex auxin response networks. (Summary by Jesus Leon @jesussaur) Nature Plants 10.1038/s41477-020-0662-y
Auxin controls virtually all facets of growth and development. This plant hormone is sensed by TIR1/AFB F-box receptors which promote ubiquitin-mediated degradation of AUX/IAA transcriptional repressors, releasing ARF transcription factors from inhibition. Phylogenetic analyses divide the ARF family into three conserved classes: A/B/C, where A-ARFs act as activators, with some B and C-ARFs acting as repressors. However, despite the apparent simplicity of this signaling module, each member belongs to gene families that are expanded in most land plants (up to 30 ARFs in Arabidopsis) and therefore may have neo- or sub- functionalized across evolution, acquiring different biochemical properties and functions, thus making the deduction of an ancestral auxin response system difficult. A recent study by Kato et al. took advantage of the liverwort Marchantia polymorpha low genetic redundancy (one TIR/AFB receptor, one AUX/IAA and three ARFs, A/B/C) to uncover the biochemical basis of a minimal auxin response system. Using a combination of molecular, structural and bioinformatic approaches, the authors deduced the M. polymorpha auxin response system, where a single A-ARF (MpARF1) switches from Aux/IAA-mediated repression to transcriptional activator in an auxin-dependent manner, whereas an auxin-insensitive B-ARF (MpARF2) antagonizes MpARF1, competing for the same target sites and acting as a repressor by recruiting the TOPLESS (TPL) corepressor. However, differential spatial distribution patterns influence the stoichiometry of these proteins, therefore resulting in the creation of zones with different auxin sensitivities to fine-tune growth and development. The consistency of this model in more complex lineages, such as Physcomitrella or Arabidopsis, suggests that this simple module may be the chassis for more complex auxin response networks. (Summary by Jesus Leon @jesussaur) Nature Plants 10.1038/s41477-020-0662-y
Plant synthetic promoters to fine tune expression of engineered genes
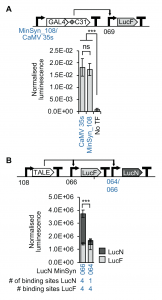 Synthetic genetic circuits allow the reconstruction of metabolic pathways in plant systems for production of many natural products including pharmaceuticals. A challenge in genetic engineering these circuits is precisely and predictably regulating gene expression, especially when genes may be desired in different ratios for optimal throughput. This research into minimal synthetic promoters (MinSyns) by Cai et al. identifies functional cis-regulatory elements that they combined to design a suite of MinSyns. Initial analysis enabled computer-guided generation of MinSyns of varying strength that are bound by endogenous transcription factors. They focused on three MinSyn systems that express in most leaf and root tissue with varying strength of which the strongest, #108, induces similar expression levels to CaMV 35S promoter. A MinSyn was used to drive expression of a synthetic transcription factor (TALE) that targets specific MinSyns co-introduced for the genes of interest. By altering the number of TALE binding sites in the reporter gene MinSyn promoters, they were able to adjust the ratio of the two proteins in the circuit. This work has exciting applications in biotechnology to improve natural product biosynthesis by optimizing enzyme ratios under the control of the same synthetic transcription factor. (Summary by Katy Dunning @plantmomkaty) bioRxiv
Synthetic genetic circuits allow the reconstruction of metabolic pathways in plant systems for production of many natural products including pharmaceuticals. A challenge in genetic engineering these circuits is precisely and predictably regulating gene expression, especially when genes may be desired in different ratios for optimal throughput. This research into minimal synthetic promoters (MinSyns) by Cai et al. identifies functional cis-regulatory elements that they combined to design a suite of MinSyns. Initial analysis enabled computer-guided generation of MinSyns of varying strength that are bound by endogenous transcription factors. They focused on three MinSyn systems that express in most leaf and root tissue with varying strength of which the strongest, #108, induces similar expression levels to CaMV 35S promoter. A MinSyn was used to drive expression of a synthetic transcription factor (TALE) that targets specific MinSyns co-introduced for the genes of interest. By altering the number of TALE binding sites in the reporter gene MinSyn promoters, they were able to adjust the ratio of the two proteins in the circuit. This work has exciting applications in biotechnology to improve natural product biosynthesis by optimizing enzyme ratios under the control of the same synthetic transcription factor. (Summary by Katy Dunning @plantmomkaty) bioRxiv
Genomic history and ecology of the geographic spread of rice
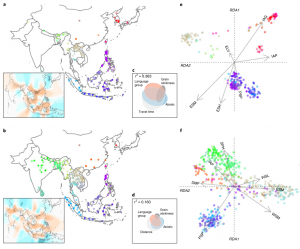 Originating in China’s Yangtze Valley ~9,000 years ago, rice (Oryza sativa) is a staple food for over half of the world’s population. While rice domestication has been well-researched, studies addressing the species’ diversification and spread post-domestication are lacking. Here, Gutaker et al. tell the “first draft of the story” of how rice was dispersed across Asia (check out this “behind the paper” piece and Twitter thread). Several fruitful collaborations facilitated an interdisciplinary modeling approach using genomic (1,400+ landrace whole-genome sequences), historical, archaeobotanical, geographical, and palaeoclimatic data. Using these methods, robust hypotheses were formed regarding the dispersal of two rice subspecies, japonica, and indica. For example, palaeoenvironmental models revealed a global cooling event ~4,200 years ago, and contemporaneously genetic data indicated a dramatic bottleneck in tropical japonica populations. A major split between temperate and tropical lineages occurred, likely resulting from some local varieties slowly adapting to northern latitudes (becoming temperate), while tropical varieties migrated to Southeast Asia where they diversified rapidly, and archaeological and historical evidence supported these patterns. Furthermore, temperature and heat accumulation gradients were identified as key environmental factors implicating genomic diversity between tropical and temperate japonica varieties. A greater understanding of how environmental factors shaped geographic distributions and evolutionary adaptations of domesticated crop species will provide valuable insights into how crops deal with imminent climatic changes, contributing to future breeding efforts and global food security. (Summary by Caroline Dowling @CarolineD0wling) Nature Plants 10.1038/s41477-020-0659-6
Originating in China’s Yangtze Valley ~9,000 years ago, rice (Oryza sativa) is a staple food for over half of the world’s population. While rice domestication has been well-researched, studies addressing the species’ diversification and spread post-domestication are lacking. Here, Gutaker et al. tell the “first draft of the story” of how rice was dispersed across Asia (check out this “behind the paper” piece and Twitter thread). Several fruitful collaborations facilitated an interdisciplinary modeling approach using genomic (1,400+ landrace whole-genome sequences), historical, archaeobotanical, geographical, and palaeoclimatic data. Using these methods, robust hypotheses were formed regarding the dispersal of two rice subspecies, japonica, and indica. For example, palaeoenvironmental models revealed a global cooling event ~4,200 years ago, and contemporaneously genetic data indicated a dramatic bottleneck in tropical japonica populations. A major split between temperate and tropical lineages occurred, likely resulting from some local varieties slowly adapting to northern latitudes (becoming temperate), while tropical varieties migrated to Southeast Asia where they diversified rapidly, and archaeological and historical evidence supported these patterns. Furthermore, temperature and heat accumulation gradients were identified as key environmental factors implicating genomic diversity between tropical and temperate japonica varieties. A greater understanding of how environmental factors shaped geographic distributions and evolutionary adaptations of domesticated crop species will provide valuable insights into how crops deal with imminent climatic changes, contributing to future breeding efforts and global food security. (Summary by Caroline Dowling @CarolineD0wling) Nature Plants 10.1038/s41477-020-0659-6
ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice
 Is it possible to simultaneously increase two incompatible features like grain yield and stress tolerance? Yin et al. suggest that optimizing cytokinin distribution in plant tissues is a promising strategy for that. ARGONAUTE2 (AGO2) and BIG GRAIN3 (BG3) genes work together promoting at the same time grain size and salt tolerance in rice. While AGO2 protein regulates chromatin condensation activating BG3 expression in AGO2-overexpressing (A2OX) plants, BG3 modulates cytokinin distribution, thereby increasing grain size and salt tolerance. The authors observed an increase in cytokinin levels in roots but a decrease in shoots, where BG3 was activated. Moreover, the same cytokinin distribution pattern was found in salt-treated plants, indicating that plants adjust this pattern to adapt to salt stress. These findings can help maintaining stable and high rice yields under saline soils. (Summary by Elisandra Pradella @Elisandra_MP) Plant Cell 10.1105/tpc.19.00542
Is it possible to simultaneously increase two incompatible features like grain yield and stress tolerance? Yin et al. suggest that optimizing cytokinin distribution in plant tissues is a promising strategy for that. ARGONAUTE2 (AGO2) and BIG GRAIN3 (BG3) genes work together promoting at the same time grain size and salt tolerance in rice. While AGO2 protein regulates chromatin condensation activating BG3 expression in AGO2-overexpressing (A2OX) plants, BG3 modulates cytokinin distribution, thereby increasing grain size and salt tolerance. The authors observed an increase in cytokinin levels in roots but a decrease in shoots, where BG3 was activated. Moreover, the same cytokinin distribution pattern was found in salt-treated plants, indicating that plants adjust this pattern to adapt to salt stress. These findings can help maintaining stable and high rice yields under saline soils. (Summary by Elisandra Pradella @Elisandra_MP) Plant Cell 10.1105/tpc.19.00542
How does fire affect germination of grasses in the Cerrado? ($)
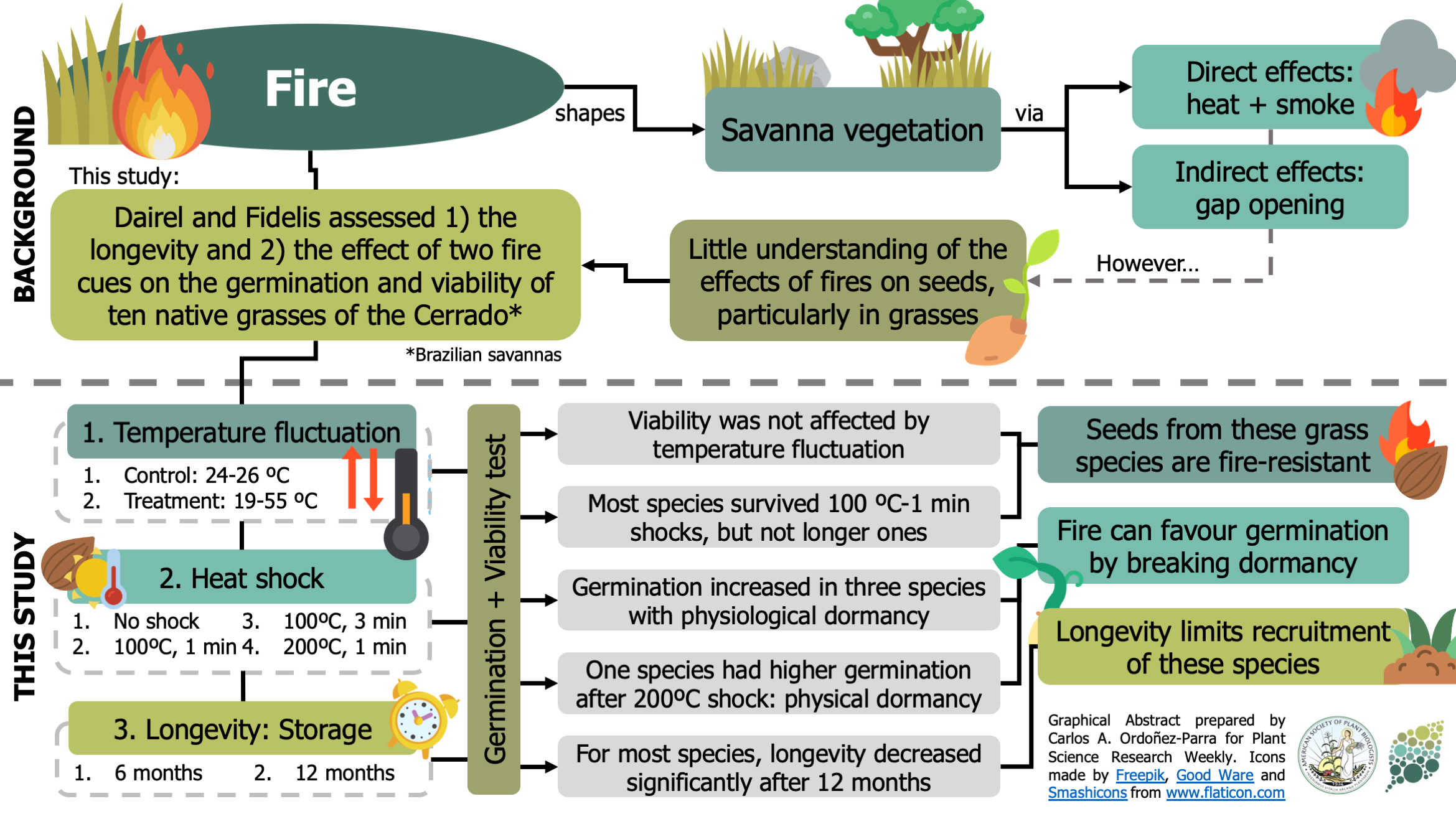 Fire shapes tropical savannas through direct (i.e., heat and smoke) and indirect (i.e., opening gaps) effects on the vegetation. However, its effects on seeds from neotropical grasses have seldom been addressed. In this paper, Dariel and Fidelis assessed the longevity and the impact of fluctuating temperatures –a consequence of gap-opening– and heat shocks on the germination and viability of ten native grass species from Brazilian savannas. The authors found that fluctuating temperatures did not affect seed viability in any species. Instead, it favored the germination of the three species with physiological dormancy. They also show that most species were able to survive heat shocks of one minute in 100ºC, but no higher temperatures or longer exposures. Surprisingly, they found two species whose germination and viability remained unaffected by heat shocks and one whose germination even peaked after exposure to 200ºC for 1 minute. With these results, the authors show that the seeds of these native grasses are resistant to fire and can even be favored by it. However, they found that most seeds had short longevity, suggesting this could be the limiting factor for the recruitment of this species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000094
Fire shapes tropical savannas through direct (i.e., heat and smoke) and indirect (i.e., opening gaps) effects on the vegetation. However, its effects on seeds from neotropical grasses have seldom been addressed. In this paper, Dariel and Fidelis assessed the longevity and the impact of fluctuating temperatures –a consequence of gap-opening– and heat shocks on the germination and viability of ten native grass species from Brazilian savannas. The authors found that fluctuating temperatures did not affect seed viability in any species. Instead, it favored the germination of the three species with physiological dormancy. They also show that most species were able to survive heat shocks of one minute in 100ºC, but no higher temperatures or longer exposures. Surprisingly, they found two species whose germination and viability remained unaffected by heat shocks and one whose germination even peaked after exposure to 200ºC for 1 minute. With these results, the authors show that the seeds of these native grasses are resistant to fire and can even be favored by it. However, they found that most seeds had short longevity, suggesting this could be the limiting factor for the recruitment of this species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000094
Shedding light on plant immunity: Light-regulated defense against P. infestans
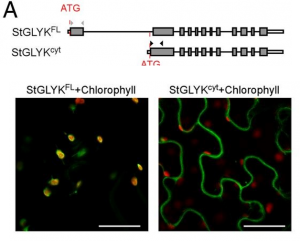 While infection by pathogens and the reciprocal immune responses are well studied in plants, the influence of other abiotic factors on these processes is not very clear. In an attempt to understand the role of light on plant defense, Gao and colleagues have shown that the AVRvnt1 effector protein secreted by Phytophthora infestans gives rise to light-dependent responses mediated by the host protein Rpi-vnt1.1 in Solanum venturii. Further analyses have revealed that these responses depend on the interaction of AVRvnt1 with chloroplast-localized glycerate 3-kinase (GLYK), which requires an N-terminal transit peptide. Interestingly, the light dependency is conferred by the light-dependent alternative promoter selection (APS) of GLYK that produces a full-length, chloroplast-localized protein in the presence of light, but in prolonged darkness produces a truncated, cytosolic protein that doesn’t interact with AVRvnt1. Also, the effector protein subverts GLYK activity by interfering with its trafficking and enabling its degradation through proteasome. This work demonstrates that Rpi-vnt1.1–mediated disease resistance is light-dependent, shedding on the regulation of immune response to a commercially important crop pest. (Summary by Pavithran Narayanan @pavi_narayanan). Proc. Natl. Acad. Sci. USA.
While infection by pathogens and the reciprocal immune responses are well studied in plants, the influence of other abiotic factors on these processes is not very clear. In an attempt to understand the role of light on plant defense, Gao and colleagues have shown that the AVRvnt1 effector protein secreted by Phytophthora infestans gives rise to light-dependent responses mediated by the host protein Rpi-vnt1.1 in Solanum venturii. Further analyses have revealed that these responses depend on the interaction of AVRvnt1 with chloroplast-localized glycerate 3-kinase (GLYK), which requires an N-terminal transit peptide. Interestingly, the light dependency is conferred by the light-dependent alternative promoter selection (APS) of GLYK that produces a full-length, chloroplast-localized protein in the presence of light, but in prolonged darkness produces a truncated, cytosolic protein that doesn’t interact with AVRvnt1. Also, the effector protein subverts GLYK activity by interfering with its trafficking and enabling its degradation through proteasome. This work demonstrates that Rpi-vnt1.1–mediated disease resistance is light-dependent, shedding on the regulation of immune response to a commercially important crop pest. (Summary by Pavithran Narayanan @pavi_narayanan). Proc. Natl. Acad. Sci. USA.
Hungry bumblebees bite plants to accelerate flowering
 Bumblebees receive nutrition from flowers in exchange for aiding plant fertilization. This mutualistic relationship relies on timely availability of flowers for the pollinators, but annual variation in flowering time in changing environments can pose a challenge. Pashalidou and Lambert et al. now found that bumblebees can actively control the flowering of plants. The authors observed that commercial bumblebee (Bombus terrestris) workers use their mandibles and tongue to nibble holes in plant leaves. Laboratory experiments showed that bee-damaged plants bear flowers about 25-30 days earlier than undamaged or mechanically damaged plants, and the damaging behavior of B. terrestris is strongly induced when pollen availability is limited. The latter observation was confirmed by semi-natural experiments conducted over two years. Two wild bumblebee species also showed leaf damaging behavior stimulated by low pollen availability, suggesting the behavior may be widespread. These findings suggest that damaging plant leaves when floral resources run short might be an adaptive trait for bumblebees to accelerate flowering. This study opens many questions, including ones important for basic plant biology: 1) “how do plants accelerate flowering upon leaf damage caused by bumblebees?”, 2) “is this an adaptive strategy for plants to increase the chance for pollination?”, and 3) “how have such mechanisms co-evolved?” (Summary by Tatsuya Nobori @nobolly) Science 10.1126/science.aay0496
Bumblebees receive nutrition from flowers in exchange for aiding plant fertilization. This mutualistic relationship relies on timely availability of flowers for the pollinators, but annual variation in flowering time in changing environments can pose a challenge. Pashalidou and Lambert et al. now found that bumblebees can actively control the flowering of plants. The authors observed that commercial bumblebee (Bombus terrestris) workers use their mandibles and tongue to nibble holes in plant leaves. Laboratory experiments showed that bee-damaged plants bear flowers about 25-30 days earlier than undamaged or mechanically damaged plants, and the damaging behavior of B. terrestris is strongly induced when pollen availability is limited. The latter observation was confirmed by semi-natural experiments conducted over two years. Two wild bumblebee species also showed leaf damaging behavior stimulated by low pollen availability, suggesting the behavior may be widespread. These findings suggest that damaging plant leaves when floral resources run short might be an adaptive trait for bumblebees to accelerate flowering. This study opens many questions, including ones important for basic plant biology: 1) “how do plants accelerate flowering upon leaf damage caused by bumblebees?”, 2) “is this an adaptive strategy for plants to increase the chance for pollination?”, and 3) “how have such mechanisms co-evolved?” (Summary by Tatsuya Nobori @nobolly) Science 10.1126/science.aay0496
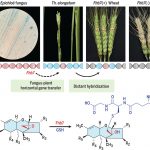
Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat
 Mycotoxins are fungal toxins with harmful health effects on humans and other animals. Fusarium head blight is a fungal disease of wheat inflorescences that can contaminate the grain and harm its consumers. Previously, Fhb7 was identified in the wheat relative Thinopyrum elongatum as a quantitative trait locus that confers resistance to Fusarium species. Here, Wang et al. cloned Fhb7. They found that this gene shows little homology to plant genes, but high homology to some fungal genes. The authors surmise that it found its way into the plant genome through horizontal gene transfer from an endophytic, often mutualistic, Epichloë fungus, some of which bioprotect against pathogenic fungi. The authors also showed that Fhb7 encodes a glutathione S-transferase that can enzymatically detoxify the fungal toxin, which is the basis for the plant’s resistance to the pathogenic fungus. Introduction of the Fhb7 gene into diverse wheat cultivars confers resistance without yield penalty. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.aba5435. See also this Perspective in the same issue by Wulff and Jones 10.1126/science.abb9991
Mycotoxins are fungal toxins with harmful health effects on humans and other animals. Fusarium head blight is a fungal disease of wheat inflorescences that can contaminate the grain and harm its consumers. Previously, Fhb7 was identified in the wheat relative Thinopyrum elongatum as a quantitative trait locus that confers resistance to Fusarium species. Here, Wang et al. cloned Fhb7. They found that this gene shows little homology to plant genes, but high homology to some fungal genes. The authors surmise that it found its way into the plant genome through horizontal gene transfer from an endophytic, often mutualistic, Epichloë fungus, some of which bioprotect against pathogenic fungi. The authors also showed that Fhb7 encodes a glutathione S-transferase that can enzymatically detoxify the fungal toxin, which is the basis for the plant’s resistance to the pathogenic fungus. Introduction of the Fhb7 gene into diverse wheat cultivars confers resistance without yield penalty. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.aba5435. See also this Perspective in the same issue by Wulff and Jones 10.1126/science.abb9991