Plant Science Research Weekly: May 1st
Review: Evolution of virulence in rust fungi — multiple solutions to one problem
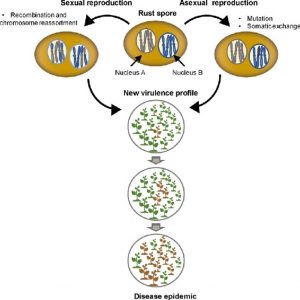 Rust fungi are a diverse group (more than 7800 species) of phytopathogenic fungi that cause considerable economic loss. (Coincidently, I’m writing on Robigalia, the Roman “anti-rust” festival, which dates from before we understood that microbes, not gods, cause disease). Figurero et al. have written an excellent update on our understanding of virulence in these fungi, which is constantly evolving to evade plant defenses; Flor’s famous gene-for-gene model of plant resistance (R gene) matching fungal gene (Avr effector gene) was developed from rust. How do rust stay so elusive? For one thing, they’re incredible complex, “with up to five distinct spore types corresponding to different life stages and with different ploidy levels,” one of which is dikaryotic, meaning it carries two different haploid nuclei within a single cell. These traits provide the opportunity for great evolutionary plasticity, with novel genetic combinations occurring both sexually and asexually, renders genomic and genetic analyses more complicated and interesting, and provides the fungus with many opportunities to evade plant defenses. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.02.007
Rust fungi are a diverse group (more than 7800 species) of phytopathogenic fungi that cause considerable economic loss. (Coincidently, I’m writing on Robigalia, the Roman “anti-rust” festival, which dates from before we understood that microbes, not gods, cause disease). Figurero et al. have written an excellent update on our understanding of virulence in these fungi, which is constantly evolving to evade plant defenses; Flor’s famous gene-for-gene model of plant resistance (R gene) matching fungal gene (Avr effector gene) was developed from rust. How do rust stay so elusive? For one thing, they’re incredible complex, “with up to five distinct spore types corresponding to different life stages and with different ploidy levels,” one of which is dikaryotic, meaning it carries two different haploid nuclei within a single cell. These traits provide the opportunity for great evolutionary plasticity, with novel genetic combinations occurring both sexually and asexually, renders genomic and genetic analyses more complicated and interesting, and provides the fungus with many opportunities to evade plant defenses. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.02.007
Review: Plant science’s next top models
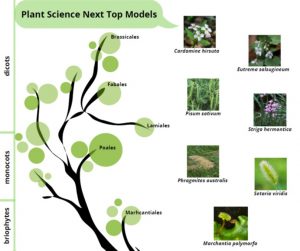 Thale cress (Arabidopsis thaliana) has been the absolute star of plant science research for more than 40 years, being the ideal model organism for its ease of handling and transferable knowledge to crops. In this review, Cesarino and coworkers explain how advances in “omics” technologies, together with improvements in molecular techniques and genetic transformation, have facilitated the use of non-model species to explore different aspects of plant development, physiology, metabolism and defense response. Across the green tree of life, the next top models and their purposes are:
Thale cress (Arabidopsis thaliana) has been the absolute star of plant science research for more than 40 years, being the ideal model organism for its ease of handling and transferable knowledge to crops. In this review, Cesarino and coworkers explain how advances in “omics” technologies, together with improvements in molecular techniques and genetic transformation, have facilitated the use of non-model species to explore different aspects of plant development, physiology, metabolism and defense response. Across the green tree of life, the next top models and their purposes are:
- Common liverwort (Marchantia polymorpha), to elucidate the origin and evolution of vascular diploid plants from basal haploid land plants
- Weeds: common reed (Phragmites australis), to determine genetic factors governing outcompetition of invasive versus native plants, and the rice parasite witchweed (Striga hermonthica) to decipher the transition from autotrophic to heterotropic life
- Close relatives of Arabidopsis: hairy bittercress (Cardamine hirsuta) to investigate the genetic basis of morphological variability, and salt cress (Eutrema salsugineum) to identify candidate genes involved in adaptation to saline environment
- Green foxtail (Setaria viridis, the wild ancestor of foxtail millet), to discover genes required for C4 photosynthesis
- Last not least, the great return of Pea (Pisum sativum): chosen by Mendel for pioneering genetic studies on inheritance, nowadays this seed crop is a model legume to study nitrogen fixation in root nodules.
Considering the enormous biodiversity in the plant kingdom, the exploitation of non-model species represents not only challenges but also opportunities for basic and applied research, towards a more sustainable agriculture relying on improved water use efficiency, reduced use of chemicals (herbicides, fertilisers) and reduced yield losses. Is this the Sunset Boulevard for the diva Arabidopsis? (Summary by Michela Osnato @michela_osnato) Ann. Bot. 10.1093/aob/mcaa063
No-Genome-Required-GWAS: Identifying genetic variants underlying phenotypic variation in plants without complete genomes
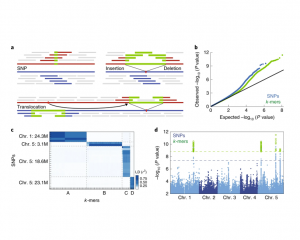 Conventional approaches to connect phenotype to genotype, including genome-wide association studies (GWAS), are often limited by the quality of the species’ reference genome, and frequently neglect to detect structural variants that are common in plant genomes. Here, Voichek and Weigel present a “No-Genome-Required-GWAS” approach (summarized in this Twitter thread), which is essentially the reverse of a conventional GWAS, that avoids using a reference genome and single nucleotide polymorphism (SNP) data. Instead, short sequences called k-mers (derived from raw sequencing data) are associated with a phenotype, and only thereafter the sequence’s genomic context is uncovered. As a proof of principle, the authors conducted a k-mer-based GWAS for flowering time in Arabidopsis, that identified associations previously detected by SNPs. The authors re-analyzed 2,000 traits in Arabidopsis, maize, and tomato, with case studies demonstrating the advantages of the k-mer-based approach versus that of SNPs. In all species, top k-mer associations were typically stronger than top SNPs. The k-mer-based approach also revealed new associations with structural variants and with regions absent from reference genomes. Moreover, k-mers successfully estimated kinship between individuals, meaning this method can be applied to species lacking high-quality reference genomes. This study emphasizes the merits of a reference genome-independent approach to detect new genetic variants, even in thoroughly studied plant species. The No-Genome-Required-GWAS has exciting relevance for linking phenotype to genotype in both model and non-model species alike. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0612-7
Conventional approaches to connect phenotype to genotype, including genome-wide association studies (GWAS), are often limited by the quality of the species’ reference genome, and frequently neglect to detect structural variants that are common in plant genomes. Here, Voichek and Weigel present a “No-Genome-Required-GWAS” approach (summarized in this Twitter thread), which is essentially the reverse of a conventional GWAS, that avoids using a reference genome and single nucleotide polymorphism (SNP) data. Instead, short sequences called k-mers (derived from raw sequencing data) are associated with a phenotype, and only thereafter the sequence’s genomic context is uncovered. As a proof of principle, the authors conducted a k-mer-based GWAS for flowering time in Arabidopsis, that identified associations previously detected by SNPs. The authors re-analyzed 2,000 traits in Arabidopsis, maize, and tomato, with case studies demonstrating the advantages of the k-mer-based approach versus that of SNPs. In all species, top k-mer associations were typically stronger than top SNPs. The k-mer-based approach also revealed new associations with structural variants and with regions absent from reference genomes. Moreover, k-mers successfully estimated kinship between individuals, meaning this method can be applied to species lacking high-quality reference genomes. This study emphasizes the merits of a reference genome-independent approach to detect new genetic variants, even in thoroughly studied plant species. The No-Genome-Required-GWAS has exciting relevance for linking phenotype to genotype in both model and non-model species alike. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0612-7
A close-up view of the thylakoids
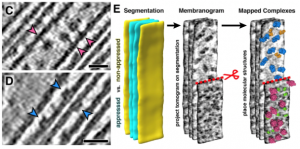 The thylakoid membranes inside the chloroplasts house the major protein complexes required for photosynthesis, including photosystems I and II (PSI/II), the b6f complex and ATP synthase. To optimize photosynthetic efficiency, the distribution and abundance of these complexes are dynamically regulated in response to varying light and growth conditions. However, to study how the spatial organization and composition of thylakoids contributes to photosynthetic productivity is challenging. Here, Wietrzynski et al. introduce “membranograms”, a cryo-EM-based method for mapping protein complexes onto segmented thylakoid membranes. This approach, here used on the green alga Chlamydomonas, gives an in situ, molecular-scale view of thylakoid architecture. Thylakoids fall into two distinct categories: appressed and non-appressed. Appressed membranes are stacked closely (~ 3 nanometer) together, whereas non-appressed ones are farther apart, and this structural distinction is a major determinant for which protein complexes are present. For example, the authors find that PSII is entirely confined to the appressed regions, whereas PSI and the ATP synthase occupies the non-appressed regions, with essentially no “mixing” at the borders. In contrast, the b6f complex, which is an important intermediary between PSII and PSI is more evenly distributed throughout the thylakoids. It will be interesting to see how this, and other properties of the thylakoid network discussed in the paper, changes when cells are challenged with different light conditions. (Summary by Frej Tulin @FrejTulin) eLIFE 10.7554/eLife.53740
The thylakoid membranes inside the chloroplasts house the major protein complexes required for photosynthesis, including photosystems I and II (PSI/II), the b6f complex and ATP synthase. To optimize photosynthetic efficiency, the distribution and abundance of these complexes are dynamically regulated in response to varying light and growth conditions. However, to study how the spatial organization and composition of thylakoids contributes to photosynthetic productivity is challenging. Here, Wietrzynski et al. introduce “membranograms”, a cryo-EM-based method for mapping protein complexes onto segmented thylakoid membranes. This approach, here used on the green alga Chlamydomonas, gives an in situ, molecular-scale view of thylakoid architecture. Thylakoids fall into two distinct categories: appressed and non-appressed. Appressed membranes are stacked closely (~ 3 nanometer) together, whereas non-appressed ones are farther apart, and this structural distinction is a major determinant for which protein complexes are present. For example, the authors find that PSII is entirely confined to the appressed regions, whereas PSI and the ATP synthase occupies the non-appressed regions, with essentially no “mixing” at the borders. In contrast, the b6f complex, which is an important intermediary between PSII and PSI is more evenly distributed throughout the thylakoids. It will be interesting to see how this, and other properties of the thylakoid network discussed in the paper, changes when cells are challenged with different light conditions. (Summary by Frej Tulin @FrejTulin) eLIFE 10.7554/eLife.53740
Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield
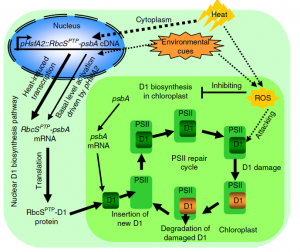 Photosystem II (PSII) is a protein complex located in the thylakoid membrane of chloroplasts that is involved in executing the initial reaction of photosynthesis in plants. When plants are exposed to extreme temperature conditions, PSII gets damaged. To repair the damage, one of the core proteins of PSII, the D1 protein, needs to be synthesized and replaced. We know that the D1 protein is synthesized from psbA messenger RNA in the chloroplast, but what will happen if the D1 protein synthesis pathway is supplemented through a gene of nuclear origin? To answer this question, Chen et al. used a bioengineering strategy to create mutant lines expressing psbA cDNA in the nucleus using the heat shock transcription factor (AtHSFA2) promoter. When these transgenic lines were grown in the presence of heat stress, they showed enhanced photosynthetic capacity and thermotolerance. Not only this, but these lines also showed increased biomass and overall yield compared to the wildtype in both control and heat stress conditions. This paper demonstrated that supplementation of D1 protein from the nucleus not only increases the repair mechanism of PSII but also leads to vigorous growth and increased net photosynthetic assimilation rates, a strategy that would be useful in addressing the impacts of global warming on plant growth and yield. (Summary by Sunita Pathak @psunita980) Nature Plants 10.1038/s41477-020-0629-z
Photosystem II (PSII) is a protein complex located in the thylakoid membrane of chloroplasts that is involved in executing the initial reaction of photosynthesis in plants. When plants are exposed to extreme temperature conditions, PSII gets damaged. To repair the damage, one of the core proteins of PSII, the D1 protein, needs to be synthesized and replaced. We know that the D1 protein is synthesized from psbA messenger RNA in the chloroplast, but what will happen if the D1 protein synthesis pathway is supplemented through a gene of nuclear origin? To answer this question, Chen et al. used a bioengineering strategy to create mutant lines expressing psbA cDNA in the nucleus using the heat shock transcription factor (AtHSFA2) promoter. When these transgenic lines were grown in the presence of heat stress, they showed enhanced photosynthetic capacity and thermotolerance. Not only this, but these lines also showed increased biomass and overall yield compared to the wildtype in both control and heat stress conditions. This paper demonstrated that supplementation of D1 protein from the nucleus not only increases the repair mechanism of PSII but also leads to vigorous growth and increased net photosynthetic assimilation rates, a strategy that would be useful in addressing the impacts of global warming on plant growth and yield. (Summary by Sunita Pathak @psunita980) Nature Plants 10.1038/s41477-020-0629-z
Nitrate defines shoot size through compensatory roles for endoreplication and cell division in Arabidopsis thaliana
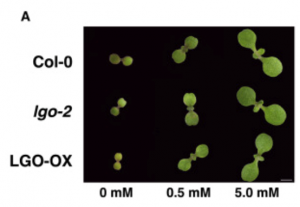 In this paper, Moreno et al. investigate how nitrate affects the balance between cell proliferation and cell expansion in shoots during early seedling development. They note that the cells in Arabidopsis cotyledons undergo considerable enlargement, and that the increase in cell size is correlated with a switch from the mitotic cell cycle to endoreplication. During endoreplication, DNA synthesis is not followed by mitosis (nuclear division), which leads to an increase in nuclear DNA. Molecularly, the mitosis/endoreplication switch is regulated by mitosis-promoting cyclin-dependent kinases (CDKs). The authors found that the CDK inhibitor LGO was upregulated in response to nitrate availability, suggesting a link between nitrogen sensing and cell cycle regulation. Consistent with this idea, mutational inactivation of LGO caused an increase in cell proliferation in nitrate-replete conditions where normally only cell expansion was observed. Interestingly, the increase in cell number compensated for their smaller size so that the final cotyledon/leaf area was unaffected, suggesting some kind of organ-level size-control mechanism. Plants generally have a rich repertoire of CDK-inhibitory proteins. Working out where and under what conditions these inhibitors act is a necessary step in understanding how the cell cycle responds changing environmental inputs. (Summary by Frej Tulin @FrejTulin) Curr. Biol. 10.1016/j.cub.2020.03.036
In this paper, Moreno et al. investigate how nitrate affects the balance between cell proliferation and cell expansion in shoots during early seedling development. They note that the cells in Arabidopsis cotyledons undergo considerable enlargement, and that the increase in cell size is correlated with a switch from the mitotic cell cycle to endoreplication. During endoreplication, DNA synthesis is not followed by mitosis (nuclear division), which leads to an increase in nuclear DNA. Molecularly, the mitosis/endoreplication switch is regulated by mitosis-promoting cyclin-dependent kinases (CDKs). The authors found that the CDK inhibitor LGO was upregulated in response to nitrate availability, suggesting a link between nitrogen sensing and cell cycle regulation. Consistent with this idea, mutational inactivation of LGO caused an increase in cell proliferation in nitrate-replete conditions where normally only cell expansion was observed. Interestingly, the increase in cell number compensated for their smaller size so that the final cotyledon/leaf area was unaffected, suggesting some kind of organ-level size-control mechanism. Plants generally have a rich repertoire of CDK-inhibitory proteins. Working out where and under what conditions these inhibitors act is a necessary step in understanding how the cell cycle responds changing environmental inputs. (Summary by Frej Tulin @FrejTulin) Curr. Biol. 10.1016/j.cub.2020.03.036
Symplastic auxin transport: Active, passive or both?
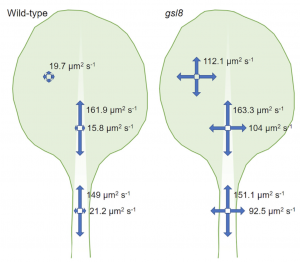 Adjacent cells in plants are connected by cytoplasmic connections called plasmodesmata (PD) and the movement through PD is called symplastic movement. Studies have shown that auxin transport between cells is protein-dependent, but it is not clear whether it can also move freely in the symplastic pathway. In this report Gao et al. show evidence for PD-mediated auxin diffusion using microscopy and mathematical modelling. Using a fluorescent tracer, the authors showed that in the epidermal cells of the leaf midrib and petiole there is more diffusion in the longitudinal compared to the transverse direction. Callose deposited in the PD neck is more abundant in the longitudinal walls compared to the transverse walls, creating a channeling effect in the longitudinal direction. This observation was supported by analysis of the mutant of GSL8 (GLUCAN SYNTHASE LIKE 8) which is deficient in callose synthesis. The authors show evidence for passive auxin movement through PD in specific epidermal cells, suggesting that auxin transport is both active and passive. Further research is needed to see how these two are coordinated. (Summary by Vijaya Batthula @Vijaya_Batthula) Curr. Biol. 10.1016/j.cub.2020.03.014
Adjacent cells in plants are connected by cytoplasmic connections called plasmodesmata (PD) and the movement through PD is called symplastic movement. Studies have shown that auxin transport between cells is protein-dependent, but it is not clear whether it can also move freely in the symplastic pathway. In this report Gao et al. show evidence for PD-mediated auxin diffusion using microscopy and mathematical modelling. Using a fluorescent tracer, the authors showed that in the epidermal cells of the leaf midrib and petiole there is more diffusion in the longitudinal compared to the transverse direction. Callose deposited in the PD neck is more abundant in the longitudinal walls compared to the transverse walls, creating a channeling effect in the longitudinal direction. This observation was supported by analysis of the mutant of GSL8 (GLUCAN SYNTHASE LIKE 8) which is deficient in callose synthesis. The authors show evidence for passive auxin movement through PD in specific epidermal cells, suggesting that auxin transport is both active and passive. Further research is needed to see how these two are coordinated. (Summary by Vijaya Batthula @Vijaya_Batthula) Curr. Biol. 10.1016/j.cub.2020.03.014
Plasma membrane aquaporin members PIPs act in concert to regulate cold acclimation and freezing tolerance responses in Arabidopsis thaliana
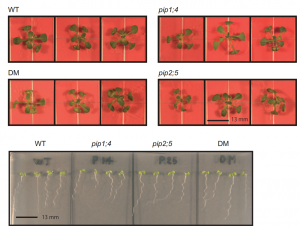 Aquaporins, also known as plasma membrane intrinsic proteins (PIP), are a large group of transporters that facilitate water transport through membranes. In this article, Rahman et al. explored the role of aquaporins in the cold stress response. They found that two aquaporin genes, PIP1;4 and PIP2;5, have increased expression in cold-acclimated plants compared to non-acclimated plants. In either pip1;4 or pip2;5 single mutant, the other gene’s expression increased, which suggests they have redundant roles. To eliminate the redundancy and elucidate the functional role of PIP1;4 and PIP2;5 in cold acclimation and freezing tolerance, authors used homozygous double mutants (pip1;4 pip2;5) and found that other aquaporin genes are up-regulated, which indicates the involvement of multiple aquaporins along with PIP1;4 and PIP2;5 in the cold responsive pathway. Single (pip1;4 and pip2;5) and double (pip1;4 pip2;5) mutants are sensitive to freezing temperature in terms of root elongation. This study illustrates that aquaporins work in concert for the cold stress response and opens an interesting research avenue. (Summary by Arif Ashraf @aribidopsis) Plant Cell Physiol. 10.1093/pcp/pcaa005
Aquaporins, also known as plasma membrane intrinsic proteins (PIP), are a large group of transporters that facilitate water transport through membranes. In this article, Rahman et al. explored the role of aquaporins in the cold stress response. They found that two aquaporin genes, PIP1;4 and PIP2;5, have increased expression in cold-acclimated plants compared to non-acclimated plants. In either pip1;4 or pip2;5 single mutant, the other gene’s expression increased, which suggests they have redundant roles. To eliminate the redundancy and elucidate the functional role of PIP1;4 and PIP2;5 in cold acclimation and freezing tolerance, authors used homozygous double mutants (pip1;4 pip2;5) and found that other aquaporin genes are up-regulated, which indicates the involvement of multiple aquaporins along with PIP1;4 and PIP2;5 in the cold responsive pathway. Single (pip1;4 and pip2;5) and double (pip1;4 pip2;5) mutants are sensitive to freezing temperature in terms of root elongation. This study illustrates that aquaporins work in concert for the cold stress response and opens an interesting research avenue. (Summary by Arif Ashraf @aribidopsis) Plant Cell Physiol. 10.1093/pcp/pcaa005
Phylogenomic evidence for reductive evolution of stomata
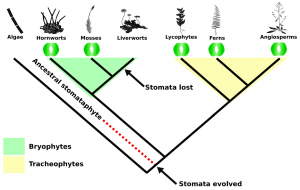 Colonization of the terrestrial environment by land plants (embryophytes), a monophyletic clade that evolved from freshwater streptophyte algae, forever changed Earth by transforming biogeochemical cycles. The evolution of stomata was a key adaptation that allowed the colonization of terra firma. Present in most land plants, stomata control the passage of carbon dioxide and water vapor, thus providing plants with a coping mechanism against arid and fluctuating conditions. The origin and ancestral function of stomata are obscure because of stomata-bearing and -lacking lineages and diversity in their morphology and function. By making use of new plant genomes and data from the 1KP project, Harris et al. sought to unravel the phylogeny of land plant and through it resolve the evolutionary history of stomatal development and function. By analyzing single-copy orthologs of 162 Viridiplantae genomes the authors determined that bryophytes were monophyletic and sister to tracheophytes, suggesting that the common ancestor of land plants already possessed stomata, with secondary loss of these in liverworts and some mosses. Subsequent presence/absence analyses of genes involved in stomata development and function confirmed this, as several genes essential for these processes were present in the common ancestor of embryophytes, whereas many of them were later lost in bryophytes. These results confirm bryophyte monophyly and shed light on stomata origin, suggesting that the ancestor of land plants possessed stomata, while bryophytes underwent reductive evolution probably due to loss of key developmental and functional regulators, with liverworts entirely losing stomata and later evolving liverwort-specific air pores. (Summary by Jesus Leon @jesussaur) Curr. Biol. 10.1016/j.cub.2020.03.048
Colonization of the terrestrial environment by land plants (embryophytes), a monophyletic clade that evolved from freshwater streptophyte algae, forever changed Earth by transforming biogeochemical cycles. The evolution of stomata was a key adaptation that allowed the colonization of terra firma. Present in most land plants, stomata control the passage of carbon dioxide and water vapor, thus providing plants with a coping mechanism against arid and fluctuating conditions. The origin and ancestral function of stomata are obscure because of stomata-bearing and -lacking lineages and diversity in their morphology and function. By making use of new plant genomes and data from the 1KP project, Harris et al. sought to unravel the phylogeny of land plant and through it resolve the evolutionary history of stomatal development and function. By analyzing single-copy orthologs of 162 Viridiplantae genomes the authors determined that bryophytes were monophyletic and sister to tracheophytes, suggesting that the common ancestor of land plants already possessed stomata, with secondary loss of these in liverworts and some mosses. Subsequent presence/absence analyses of genes involved in stomata development and function confirmed this, as several genes essential for these processes were present in the common ancestor of embryophytes, whereas many of them were later lost in bryophytes. These results confirm bryophyte monophyly and shed light on stomata origin, suggesting that the ancestor of land plants possessed stomata, while bryophytes underwent reductive evolution probably due to loss of key developmental and functional regulators, with liverworts entirely losing stomata and later evolving liverwort-specific air pores. (Summary by Jesus Leon @jesussaur) Curr. Biol. 10.1016/j.cub.2020.03.048
A plant secondary metabolite selectively targets bacterial pathogenicity
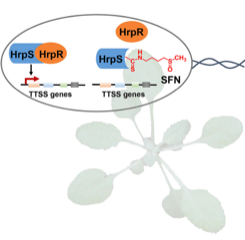 Plants have a variety of secondary metabolites that are associated with defense against pathogens, but the mode of action of such metabolites is poorly understood. Wang et al. revealed that the Arabidopsis secondary metabolite sulforaphane (SFN), which is derived from aliphatic glucosinolate, suppresses transcript accumulation of genes involved in the type III secretion system (T3SS), an essential virulence component of the bacterial pathogen Pseudomonas syringae. SFN accumulates in the leaf apoplast, site of P. syringae colonization, in a manner dependent on MYB28 and MYB29 transcription factors. In myb28myb29 mutant plants, exogenous application of SFN at the physiological concentration was sufficient to suppress T3SS expression and bacterial growth. SFN did not affect bacterial growth in vitro at the physiological concentration, and the leaf microbiota of the myb28myb29 plants grown in natural soil resembled that of wild type plants, suggesting that SFN does not function as a general antimicrobial in plants. The authors identified that SFN preferentially reacts with Cys209 of HrpS, an important regulator of the T3SS. Further, they showed that HrpSCys209 is required for HrpS to induce the T3SS and promote bacterial growth in planta. Interestingly, HrpSCys209 is highly conserved in P. syringae isolates non-adapted to Arabidopsis, but not in adapted isolates, raising a possibility that targeting of HrpS by SFN may pose a selective pressure on the evolution of plant-associated bacteria. This study provides mechanistic insights into an understudied but vital question: “how do plants affect pathogens to suppress their growth?” (Summary by Tatsuya Nobori @nobolly) Cell Host & Microbe 10.1016/j.chom.2020.03.004
Plants have a variety of secondary metabolites that are associated with defense against pathogens, but the mode of action of such metabolites is poorly understood. Wang et al. revealed that the Arabidopsis secondary metabolite sulforaphane (SFN), which is derived from aliphatic glucosinolate, suppresses transcript accumulation of genes involved in the type III secretion system (T3SS), an essential virulence component of the bacterial pathogen Pseudomonas syringae. SFN accumulates in the leaf apoplast, site of P. syringae colonization, in a manner dependent on MYB28 and MYB29 transcription factors. In myb28myb29 mutant plants, exogenous application of SFN at the physiological concentration was sufficient to suppress T3SS expression and bacterial growth. SFN did not affect bacterial growth in vitro at the physiological concentration, and the leaf microbiota of the myb28myb29 plants grown in natural soil resembled that of wild type plants, suggesting that SFN does not function as a general antimicrobial in plants. The authors identified that SFN preferentially reacts with Cys209 of HrpS, an important regulator of the T3SS. Further, they showed that HrpSCys209 is required for HrpS to induce the T3SS and promote bacterial growth in planta. Interestingly, HrpSCys209 is highly conserved in P. syringae isolates non-adapted to Arabidopsis, but not in adapted isolates, raising a possibility that targeting of HrpS by SFN may pose a selective pressure on the evolution of plant-associated bacteria. This study provides mechanistic insights into an understudied but vital question: “how do plants affect pathogens to suppress their growth?” (Summary by Tatsuya Nobori @nobolly) Cell Host & Microbe 10.1016/j.chom.2020.03.004
Interplay between phosphorylation and ubiquitination is key for regulating reactive oxygen species during plant immunity
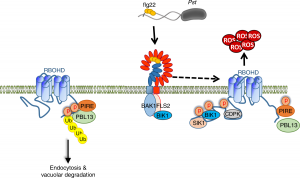 Reactive oxygen species (ROS) are essential secondary messengers for rapid transmission of local and long-distance signalling in plants. Respiratory oxidase homologs (RBOHs) are the family of enzymes in plants that produce extracellular ROS, with RBOHD being the primary oxidase in Arabidopsis for ROS production following pathogen perception. Positive regulation of RBOHD by activating phosphorylation is well characterized, but it was unknown how RHOBD is inhibited pre- and post-pathogen recognition to maintain immune homeostasis. Lee et al. identified that PBS1-like kinase 13 (PBL13) phosphorylates conserved C-terminal residues on RBOHD to decrease its stability. They further identified the previously uncharacterized ubiquitin E3 ligase PIRE (PBL13 interacting RING domain E3 ligase), which polyubiquitinates RBOHD at residues near the PBL13-targetted phosphorylation sites, targeting it for endocytosis and vacuolar degradation. Mimicking phosphorylation at these sites directly promotes ubiquitination and protein turnover. These results showcase cross-talk between phosphorylation and ubiquitination as a mechanism to negatively regulate plant immunity. (Summary by Katy Dunning @plantmomkaty) Nature Comms 10.1038/s41467-020-15601-5
Reactive oxygen species (ROS) are essential secondary messengers for rapid transmission of local and long-distance signalling in plants. Respiratory oxidase homologs (RBOHs) are the family of enzymes in plants that produce extracellular ROS, with RBOHD being the primary oxidase in Arabidopsis for ROS production following pathogen perception. Positive regulation of RBOHD by activating phosphorylation is well characterized, but it was unknown how RHOBD is inhibited pre- and post-pathogen recognition to maintain immune homeostasis. Lee et al. identified that PBS1-like kinase 13 (PBL13) phosphorylates conserved C-terminal residues on RBOHD to decrease its stability. They further identified the previously uncharacterized ubiquitin E3 ligase PIRE (PBL13 interacting RING domain E3 ligase), which polyubiquitinates RBOHD at residues near the PBL13-targetted phosphorylation sites, targeting it for endocytosis and vacuolar degradation. Mimicking phosphorylation at these sites directly promotes ubiquitination and protein turnover. These results showcase cross-talk between phosphorylation and ubiquitination as a mechanism to negatively regulate plant immunity. (Summary by Katy Dunning @plantmomkaty) Nature Comms 10.1038/s41467-020-15601-5
The morphology, molecular development and ecological function of pseudonectaries on Nigella damascena (Ranunculaceae) petals
 Pseudonectaries do not secrete nectar like nectaries. So, what are their ecological functions? Are there any morphological and anatomical differences between them and true nectaries? How do they develop and evolve? Liao et al. examined pseudonectaries of Nigella damascene to answer these questions. The epidermis of N. damascene pseudonectaries is composed of polygonal cells with smooth and grainy surfaces, which are shiny and reflective and thus attract the attention of honey bees. The development of pseudonectaries strongly coincided with the ectopic expression of the abaxial gene NidaYAB5, and knockdown of the gene resulted in the lack of pseudonectaries and less attractive flowers, significantly decreasing both the visit frequency and probing time of honey bees. Genes related to cell division, chloroplast development, and wax formation were also associated with pseudonectary formation. Overall, pseudonectaries attract suitable pollinators and also guide their visit by marking the entrance of the nectar chamber. (Summary by Elisandra Pradella @Elisandra_MP) Nature Comms. 10.1038/s41467-020-15658-2
Pseudonectaries do not secrete nectar like nectaries. So, what are their ecological functions? Are there any morphological and anatomical differences between them and true nectaries? How do they develop and evolve? Liao et al. examined pseudonectaries of Nigella damascene to answer these questions. The epidermis of N. damascene pseudonectaries is composed of polygonal cells with smooth and grainy surfaces, which are shiny and reflective and thus attract the attention of honey bees. The development of pseudonectaries strongly coincided with the ectopic expression of the abaxial gene NidaYAB5, and knockdown of the gene resulted in the lack of pseudonectaries and less attractive flowers, significantly decreasing both the visit frequency and probing time of honey bees. Genes related to cell division, chloroplast development, and wax formation were also associated with pseudonectary formation. Overall, pseudonectaries attract suitable pollinators and also guide their visit by marking the entrance of the nectar chamber. (Summary by Elisandra Pradella @Elisandra_MP) Nature Comms. 10.1038/s41467-020-15658-2
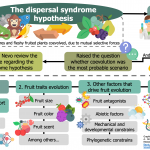
The dispersal syndrome hypothesis: How animals shaped fruit traits, and how they did not
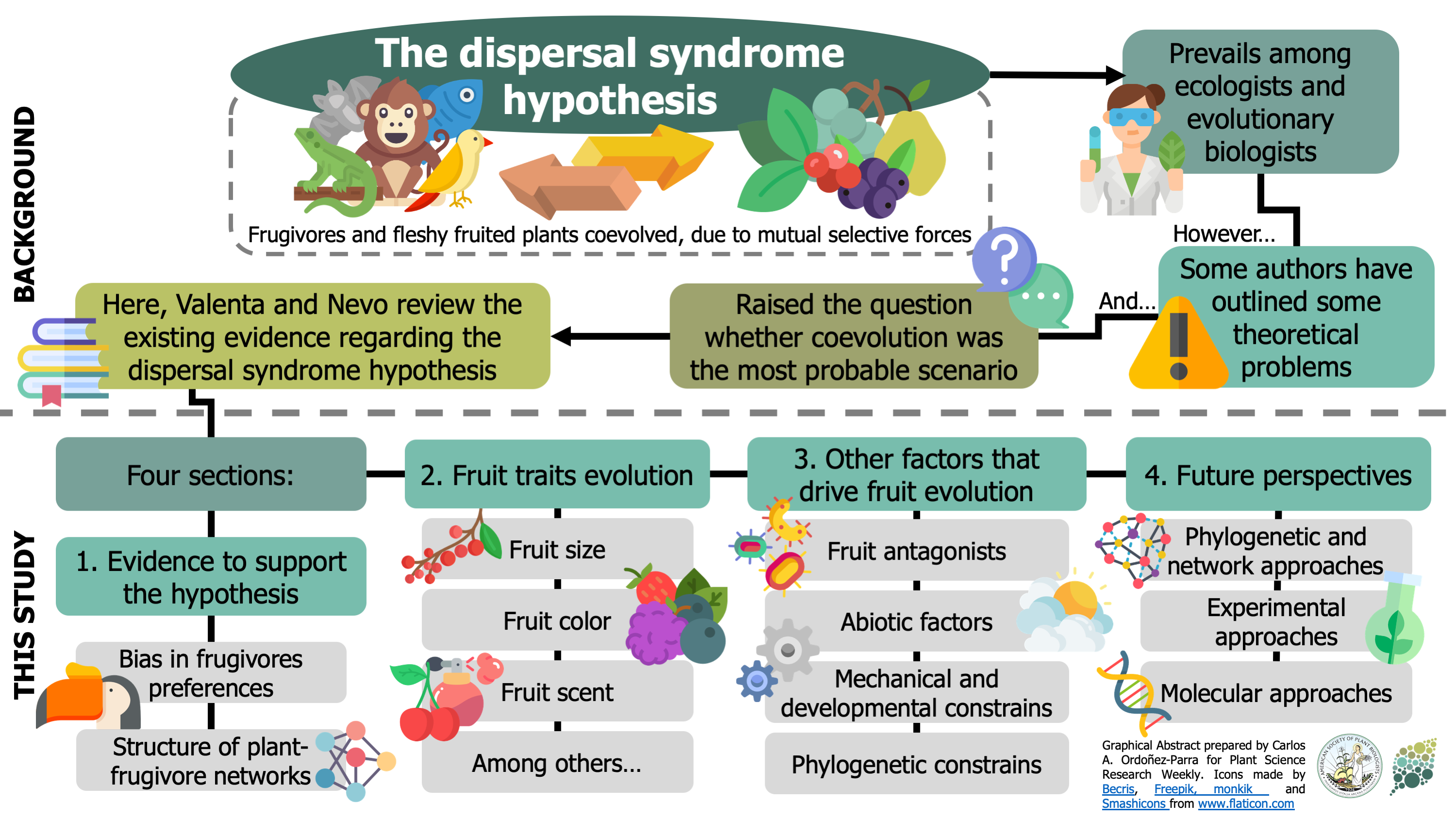 The dispersal syndrome hypothesis states that frugivores and fleshy-fruited plants coevolve by exerting selective pressures on each other. Even though this idea still prevails, some theoretical issues could make this scenario improbable. As a result, a new chicken-or-egg dilemma appeared: are the frugivores responsible for fruit evolution, or is it the opposite? In this fascinating review, Valenta & Nevo bring together evidence that either supports or conflicts with the dispersal syndrome hypothesis. First, the authors introduce the evidence confirming it, such as the bias of frugivores when selecting fruits and the non-random structure of plant-frugivore networks. For instance, they show how fruit size, color, and scent, among other traits, have been subject to frugivores’ selection. However, the authors also present other factors that could have influenced fleshy fruits evolution besides frugivores, showing this dilemma has more protagonists. In the end, the review –an obligate reading for those interested in plant-frugivore interactions– describes how future studies could contribute to the effort of revealing “the forces driving the tremendous diversity of fruit traits” (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Funct. Ecol. 10.1111/1365-2435.13564
The dispersal syndrome hypothesis states that frugivores and fleshy-fruited plants coevolve by exerting selective pressures on each other. Even though this idea still prevails, some theoretical issues could make this scenario improbable. As a result, a new chicken-or-egg dilemma appeared: are the frugivores responsible for fruit evolution, or is it the opposite? In this fascinating review, Valenta & Nevo bring together evidence that either supports or conflicts with the dispersal syndrome hypothesis. First, the authors introduce the evidence confirming it, such as the bias of frugivores when selecting fruits and the non-random structure of plant-frugivore networks. For instance, they show how fruit size, color, and scent, among other traits, have been subject to frugivores’ selection. However, the authors also present other factors that could have influenced fleshy fruits evolution besides frugivores, showing this dilemma has more protagonists. In the end, the review –an obligate reading for those interested in plant-frugivore interactions– describes how future studies could contribute to the effort of revealing “the forces driving the tremendous diversity of fruit traits” (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Funct. Ecol. 10.1111/1365-2435.13564