Plant Science Research Weekly: May 13, 2022
Review: Protein phosphorylation “toggle switch” for plant iron balance
 Protein phosphorylation and dephosphorylation act as a switch regulating a multitude of protein properties, be it their activity, interaction with other proteins, stability, or even cellular localization. This review by Li et al. describs the many ways that protein phosphorylation contributes to iron uptake and homeostasis. The membrane localization of the iron transporter IRT1 is regulated by Sorting Nexin 1 (SNX1), and phosphorylation of SNX1and IRT1 regulates iron uptake and the intracellular iron pool. When exposed to above-optimal zinc and manganese concentrations, phosphorylation by the CBL-Interacting protein kinase 23 (CIPK23) targets IRT1 for proteasomal degradation and prevents IRT1 mediated non-Fe metal transport. Iron homeostasis regulators like the bHLH family transcription factors FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) and POPEYE (PYE) are also regulated by phosphorylation. For example, phosphorylation of FIT at Ser272 by the CIPK11-CBL1/9 network renders it active while phosphorylation at Tyr 238 and Tyr 272, putatively by MAP kinases MPK3 and MPK6, shuts it down. The UPSTREAM REGULATOR OF IRT1 (URI), a modulator of FIT and PYE, is regulated by iron-status dependent phosphorylation for both activity and tissue localization. Under iron deficiency, URI is phosphorylated, and the formation of its heterodimer with bHLH TFs is promoted. The heterodimer formation of URI drives the transcription of key members of the bHLHIb TF family that are inducers of FIT dependent iron uptake responses. Finally, ethylene and nitric oxide-mediated iron deficiency responses are also immensely reliant on protein phosphorylation. Protein phosphorylation is thus an essential strategy for modulating key regulators to ensure a highly sensitive and precise response to iron availability. (Summary by Lekshmy Sathee @lekshmysnair) Trends Plant Sci. 10.1016/j.tplants.2022.03.012
Protein phosphorylation and dephosphorylation act as a switch regulating a multitude of protein properties, be it their activity, interaction with other proteins, stability, or even cellular localization. This review by Li et al. describs the many ways that protein phosphorylation contributes to iron uptake and homeostasis. The membrane localization of the iron transporter IRT1 is regulated by Sorting Nexin 1 (SNX1), and phosphorylation of SNX1and IRT1 regulates iron uptake and the intracellular iron pool. When exposed to above-optimal zinc and manganese concentrations, phosphorylation by the CBL-Interacting protein kinase 23 (CIPK23) targets IRT1 for proteasomal degradation and prevents IRT1 mediated non-Fe metal transport. Iron homeostasis regulators like the bHLH family transcription factors FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) and POPEYE (PYE) are also regulated by phosphorylation. For example, phosphorylation of FIT at Ser272 by the CIPK11-CBL1/9 network renders it active while phosphorylation at Tyr 238 and Tyr 272, putatively by MAP kinases MPK3 and MPK6, shuts it down. The UPSTREAM REGULATOR OF IRT1 (URI), a modulator of FIT and PYE, is regulated by iron-status dependent phosphorylation for both activity and tissue localization. Under iron deficiency, URI is phosphorylated, and the formation of its heterodimer with bHLH TFs is promoted. The heterodimer formation of URI drives the transcription of key members of the bHLHIb TF family that are inducers of FIT dependent iron uptake responses. Finally, ethylene and nitric oxide-mediated iron deficiency responses are also immensely reliant on protein phosphorylation. Protein phosphorylation is thus an essential strategy for modulating key regulators to ensure a highly sensitive and precise response to iron availability. (Summary by Lekshmy Sathee @lekshmysnair) Trends Plant Sci. 10.1016/j.tplants.2022.03.012
A plant-specific module for homologous recombination repair
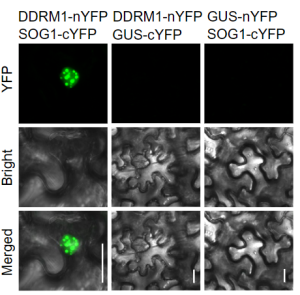 Damage to DNA occurs through exposure to environmental stresses in animal and plant kingdoms alike. While the former groups are relatively well explored when it comes to DNA damage response (DDR) mechanisms, plant-related knowledge lags behind. A recent study by Wang et al. revealed a unique plant DDR protein identified through genetic screens of the TRANSPLANTA collection containing 1,636 homozygous Arabidopsis thaliana lines. The screen identified the DNA damage response ddrm1 mutant which was sensitive to DNA damage-inducing reagents. DDRM1 is conserved in land plants and encodes a nuclear localized protein. Using split luciferase and BiFC assays, DDRM1 was confirmed to interact with the DDR master regulator transcription factor SOG1, leading to its ubiquitination. Mono-ubiquitination of SOG1 at numerous lysine residues leads to its subsequent stabilization, influencing the ability of SOG1 to mediate cell cycle arrest, DNA repair and cell death. This study enhances our understanding of plant specific DNA damage responses and posttranslational regulation of this critical DDR pathway. (Summary by Orla Sherwood, @orlasherwood) PNAS. 10.1073/pnas.2202970119
Damage to DNA occurs through exposure to environmental stresses in animal and plant kingdoms alike. While the former groups are relatively well explored when it comes to DNA damage response (DDR) mechanisms, plant-related knowledge lags behind. A recent study by Wang et al. revealed a unique plant DDR protein identified through genetic screens of the TRANSPLANTA collection containing 1,636 homozygous Arabidopsis thaliana lines. The screen identified the DNA damage response ddrm1 mutant which was sensitive to DNA damage-inducing reagents. DDRM1 is conserved in land plants and encodes a nuclear localized protein. Using split luciferase and BiFC assays, DDRM1 was confirmed to interact with the DDR master regulator transcription factor SOG1, leading to its ubiquitination. Mono-ubiquitination of SOG1 at numerous lysine residues leads to its subsequent stabilization, influencing the ability of SOG1 to mediate cell cycle arrest, DNA repair and cell death. This study enhances our understanding of plant specific DNA damage responses and posttranslational regulation of this critical DDR pathway. (Summary by Orla Sherwood, @orlasherwood) PNAS. 10.1073/pnas.2202970119
Xylan-based nanocompartments orchestrate plant vessel wall patterning
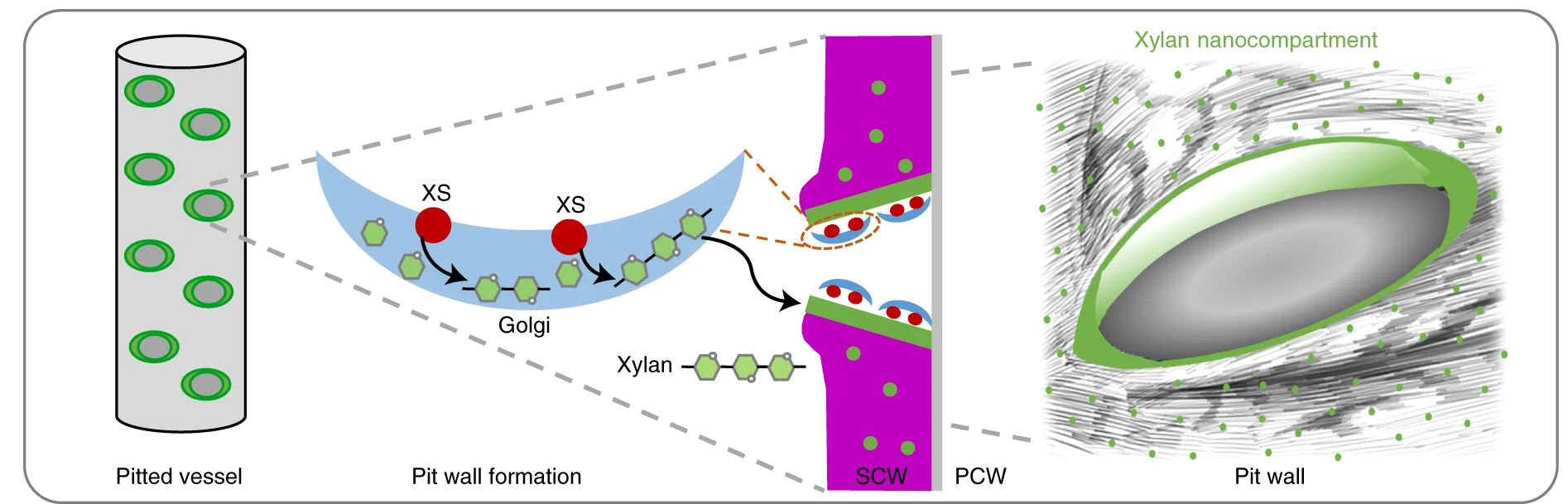 Xylem vessels have a distinct secondary cell wall (SCW) formation, where the metaxylem stage contains a characteristic pit pattern. The hemicellulose xylan is an important component of the SCW, functioning as a crosslinking polymer that coats cellulose microfibrils and interacts with other SCW components. However, the xylan initiation mechanism was unknown. Here, Wang et al. found that the IRX10 protein in rice (ortholog of a known Arabidopsis protein) and its homologues perform dual roles in xylan initiation and elongation during xylan synthesis, using in vitro enzyme studies. They demonstrate in both rice and Arabidopsis that xylan accumulates at the pit borders of the metaxylem, which is disrupted in irx10 mutants. Using various microscopy techniques, they show that the mutants have larger pit areas and smaller vessels, and reduced xylose content. Additionally, the cellulosic nanofibers in the SCW are not properly packed, and the SCW defects result in impaired xylem water transport function. (Summary by Jiawen Chen @Jiaaawen) Nature Plants 10.1038/s41477-022-01113-1.
Xylem vessels have a distinct secondary cell wall (SCW) formation, where the metaxylem stage contains a characteristic pit pattern. The hemicellulose xylan is an important component of the SCW, functioning as a crosslinking polymer that coats cellulose microfibrils and interacts with other SCW components. However, the xylan initiation mechanism was unknown. Here, Wang et al. found that the IRX10 protein in rice (ortholog of a known Arabidopsis protein) and its homologues perform dual roles in xylan initiation and elongation during xylan synthesis, using in vitro enzyme studies. They demonstrate in both rice and Arabidopsis that xylan accumulates at the pit borders of the metaxylem, which is disrupted in irx10 mutants. Using various microscopy techniques, they show that the mutants have larger pit areas and smaller vessels, and reduced xylose content. Additionally, the cellulosic nanofibers in the SCW are not properly packed, and the SCW defects result in impaired xylem water transport function. (Summary by Jiawen Chen @Jiaaawen) Nature Plants 10.1038/s41477-022-01113-1.
Cytokinin–CLAVATA cross-talk is an ancient mechanism regulating shoot meristem homeostasis in land plants
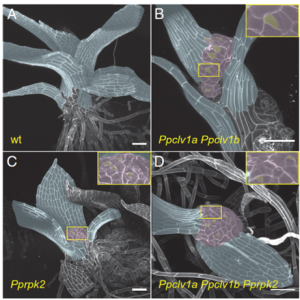 The shoot apical meristem (SAM) maintains a small pool of pluripotent stem cells, which are the source of all above-ground plant tissues. In Arabidopsis thaliana, CLAVATA3 (CLV3), CLAVATA1(CLV1) and RECEPTOR-LIKE PROTEIN KINASE2 (RPK2) repress WUSCHEL (WUS) expression. WUS induces CLV3, forming a local feedback loop to maintain the stem cell population. Cytokinin is also involved in the WUS-CLV regulation, as well as SAM formation, maintenance, and growth. In contrast to flowering plants, the moss SAM is self-generated from a single stem cell and there is no stem cell maintenance through WUS function. Since the functions of CLV1 and RPK2 are conserved in the moss Physcomitrium patens, Cammarata et al. investigated whether CLV and cytokinin cross-talk is conserved in a similar way despite lacking the WUS interaction. Exogenous cytokinin treatment, CRISPR mutants, and mathematical modeling revealed that CLV1 and RPK2 act additively in parallel pathways to regulate stem cell specification. Mutants of CLV and RPK form ectopic stem cells in response to exogenous cytokinin, supporting the hypothesis that PpCLV1 and PpRPK2 act upstream of cytokinin-stem cell induction. This work shows that CLV and cytokinin cross-talk is conserved to maintain SAM homeostasis in P. patens, demonstrates how the pathways between different plant lineages are conserved, and is a valuable contribution to our understanding of the underlying genetic interactions. (Summary by Andrea Gómez Felipe @andreagomezfe) PNAS 10.1073/pnas.2116860119
The shoot apical meristem (SAM) maintains a small pool of pluripotent stem cells, which are the source of all above-ground plant tissues. In Arabidopsis thaliana, CLAVATA3 (CLV3), CLAVATA1(CLV1) and RECEPTOR-LIKE PROTEIN KINASE2 (RPK2) repress WUSCHEL (WUS) expression. WUS induces CLV3, forming a local feedback loop to maintain the stem cell population. Cytokinin is also involved in the WUS-CLV regulation, as well as SAM formation, maintenance, and growth. In contrast to flowering plants, the moss SAM is self-generated from a single stem cell and there is no stem cell maintenance through WUS function. Since the functions of CLV1 and RPK2 are conserved in the moss Physcomitrium patens, Cammarata et al. investigated whether CLV and cytokinin cross-talk is conserved in a similar way despite lacking the WUS interaction. Exogenous cytokinin treatment, CRISPR mutants, and mathematical modeling revealed that CLV1 and RPK2 act additively in parallel pathways to regulate stem cell specification. Mutants of CLV and RPK form ectopic stem cells in response to exogenous cytokinin, supporting the hypothesis that PpCLV1 and PpRPK2 act upstream of cytokinin-stem cell induction. This work shows that CLV and cytokinin cross-talk is conserved to maintain SAM homeostasis in P. patens, demonstrates how the pathways between different plant lineages are conserved, and is a valuable contribution to our understanding of the underlying genetic interactions. (Summary by Andrea Gómez Felipe @andreagomezfe) PNAS 10.1073/pnas.2116860119
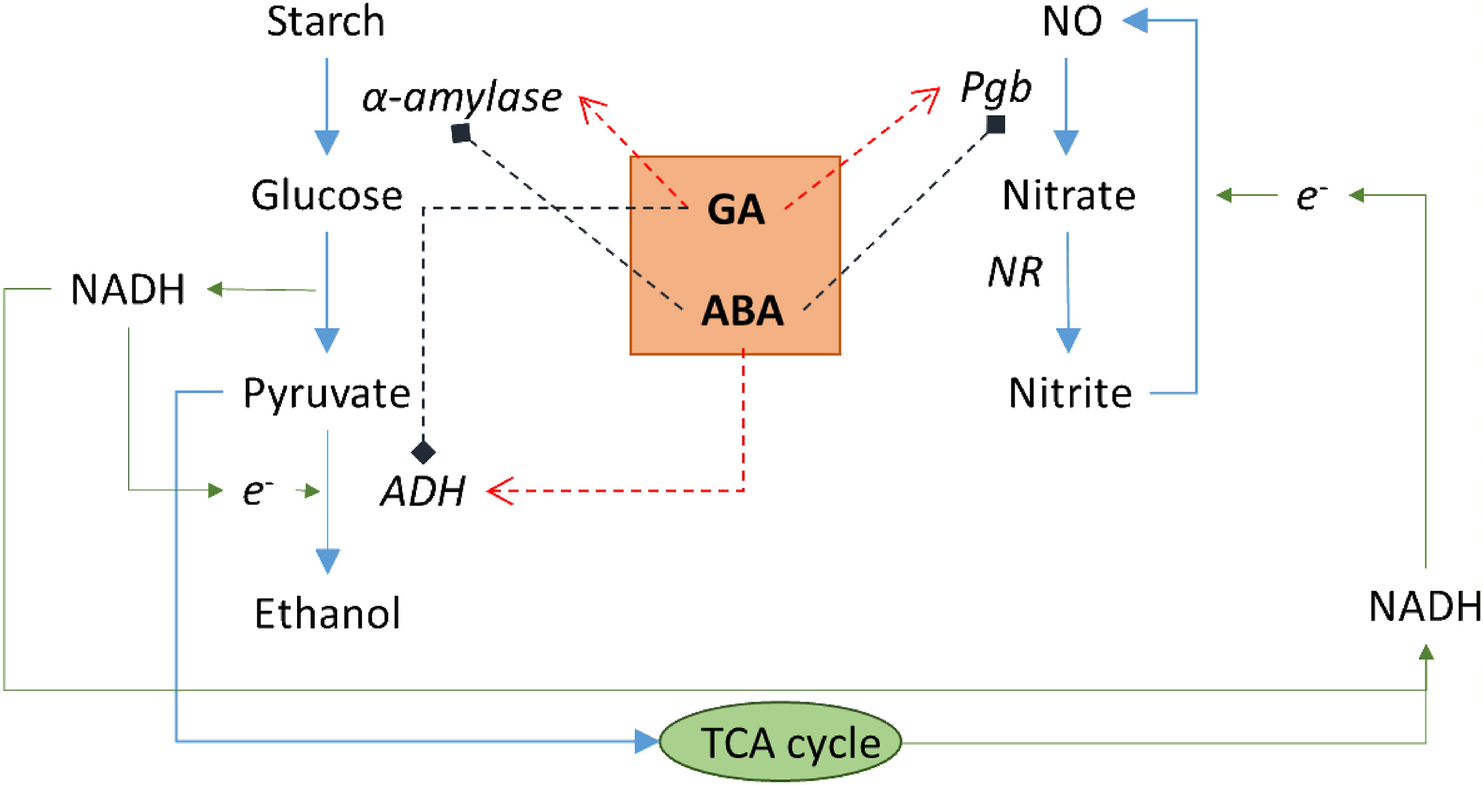
Anaerobiosis modulation of two phytoglobins in barley (Hordeum vulgare L.), and their regulation by gibberellin and abscisic acid in aleurone cells
Barley seeds are anoxic, and during germination pathways are induced to allow energy to be obtained without oxygen. Phytoglobins (Pgb) are hypoxia-induced proteins that are involved in energy metabolism during germination through the Pgb-NO cycle, an alternative pathway to fermentation. In the Pgb-NO cycle, Pgb1 scavenge NO to produce nitrate, which is converted to nitrite by nitrate reductase, and again to NO by nitrate-NO reductase, producing ATP. As in many other cereals, the germination process is tightly controlled by abscisic acid (ABA) and gibberellic acid (GA) hormones. In this work, Nie et al. studied the effect of oxygen deficiency on the amount of Pgb transcripts and proteins, the expression of fermentation-related genes such as alcohol dehydrogenase (ADH) and the activity of α-amylase. The authors conclude that Pgb induction in aleurone cells is related to anaerobiosis and that GA upregulates not only α-amylase but also Pgb transcripts. Thus, the energy demands during the first hours on the germinating seed may be satisfied by the Pgb-NO cycle. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Plant Physiol. Biochem. 10.1016/j.plaphy.2022.04.014
N-mediated heading date-1 (Nhd1) regulates nitrogen uptake efficiency and root growth in rice
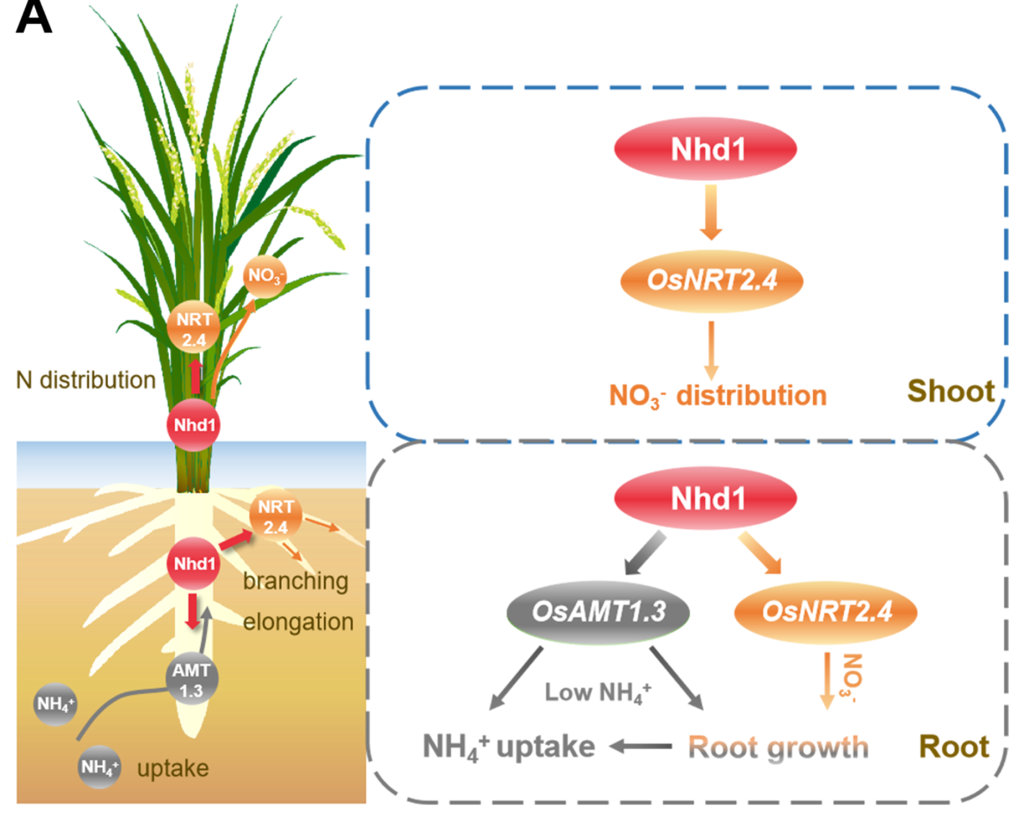 In rice, the transcription factor N-mediated heading date-1 (Nhd1) activates the expression of Heading date 3a (Hd3a), a floral regulator gene and regulate flowering. Li et al. report the role of Nhd1 in nitrogen (N) use efficiency and N-dependent root growth in rice. nhd1 mutants were impaired in root growth when grown in low nitrate, high nitrate, and in low ammonium conditions. Nhd1 induces the expression of thedual affinity nitrate transporter NRT2.4 and the putative ammonium transceptor AMT1.3 by recognising the NBS (Nhd1 binding site) motifs in the respective promoter regions. Under low ammonium supply, the rate of ammonium influx and root growth of amt1.3 and nhd1 mutants were similar. The root growth reduction, rate of nitrate uptake and impairment in nitrate translocation was similar in nrt2.4 and nhd1 mutants. Nhd1 is thus an upstream regulator of N-form dependent root growth responses controlled by NRT2.4 and AMT1.3. The over-expression of AMT1.3 and NRT2.4 could rescue impaired N uptake of nhd1 lines, without altering flowering time and HD3a expression. Interestingly, when grown until maturity under field conditions, the total N uptake was significantly improved in nhd1 lines, even when the soil N was low. The improved N uptake efficiency of nhd1 lines was probably due to the extended growth duration, and, due to delayed flowering, a more favorable temperature during the reproductive stage. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac178
In rice, the transcription factor N-mediated heading date-1 (Nhd1) activates the expression of Heading date 3a (Hd3a), a floral regulator gene and regulate flowering. Li et al. report the role of Nhd1 in nitrogen (N) use efficiency and N-dependent root growth in rice. nhd1 mutants were impaired in root growth when grown in low nitrate, high nitrate, and in low ammonium conditions. Nhd1 induces the expression of thedual affinity nitrate transporter NRT2.4 and the putative ammonium transceptor AMT1.3 by recognising the NBS (Nhd1 binding site) motifs in the respective promoter regions. Under low ammonium supply, the rate of ammonium influx and root growth of amt1.3 and nhd1 mutants were similar. The root growth reduction, rate of nitrate uptake and impairment in nitrate translocation was similar in nrt2.4 and nhd1 mutants. Nhd1 is thus an upstream regulator of N-form dependent root growth responses controlled by NRT2.4 and AMT1.3. The over-expression of AMT1.3 and NRT2.4 could rescue impaired N uptake of nhd1 lines, without altering flowering time and HD3a expression. Interestingly, when grown until maturity under field conditions, the total N uptake was significantly improved in nhd1 lines, even when the soil N was low. The improved N uptake efficiency of nhd1 lines was probably due to the extended growth duration, and, due to delayed flowering, a more favorable temperature during the reproductive stage. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac178
Machine learning algorithms predict soil seed bank persistence from easily available traits
 Knowing whether a given plant species forms a persistent soil seed bank is essential to understanding its ecological dynamics, yet soil seed bank studies are both labor-intensive and time-consuming. One could argue that models should help predict this. Still, seed persistence in the soil is shaped by multiple plant and environmental characteristics, so conventional statistical methods might not suit the task. Fortunately, Rosbakh, Pichler and Poschlod show us that machine learning approaches are up to the challenge. Their machine learning model (i.e., random forest) outperformed the traditional, generalized linear model when predicting the type (persistent vs. transient) and density (seeds/m2) of the soil seed bank. Moreover, the machine learning approach retained high predictive power with just a few easily measurable variables, such as site temperature and seed mass. In fact, it recovered several interactions between seed persistence and environmental and plant characteristics previously described in the literature. As a result, this research outlines a promising tool for increasing our knowledge about soil seed banks, especially in areas with limited resources. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Appl. Veg. Sci. 10.1111/avsc.12660
Knowing whether a given plant species forms a persistent soil seed bank is essential to understanding its ecological dynamics, yet soil seed bank studies are both labor-intensive and time-consuming. One could argue that models should help predict this. Still, seed persistence in the soil is shaped by multiple plant and environmental characteristics, so conventional statistical methods might not suit the task. Fortunately, Rosbakh, Pichler and Poschlod show us that machine learning approaches are up to the challenge. Their machine learning model (i.e., random forest) outperformed the traditional, generalized linear model when predicting the type (persistent vs. transient) and density (seeds/m2) of the soil seed bank. Moreover, the machine learning approach retained high predictive power with just a few easily measurable variables, such as site temperature and seed mass. In fact, it recovered several interactions between seed persistence and environmental and plant characteristics previously described in the literature. As a result, this research outlines a promising tool for increasing our knowledge about soil seed banks, especially in areas with limited resources. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Appl. Veg. Sci. 10.1111/avsc.12660
Fungal exopolysaccharide regulates plant-microbe interaction
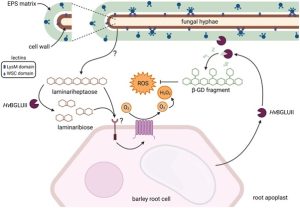 The studies on the apoplastic interactions of plants and fungi often focus on the fungal cell-wall, but recent studies indicate various plant colonizing fungi also have an exopolysaccharide (EPS) layer outside their cell wall. Chandrasekar et al. observed that the composition of this fungal EPS is distinct from the fungal cell wall and the secreted culture filtrate, indicating it is neither just an extension of the cell wall nor something simply secreted out into the medium. The activity of a barley glucanase (which is secreted in response to fungal treatment) on the EPS that is secreted by endophytic or pathogenic fungi leads to the release of a β-glucan decasaccharide (β-GD). Interestingly application of β-GD to barley or Arabidopsis suppresses the immune-responsive ROS burst triggered by other elicitors of immunity, but it does not affect other early immune responses. In vitro studies indicate β-GD can scavenge ROS indicating it might be quenching apoplastic ROS that is secreted by plants during plant-microbe interactions. Co-treatment of plants with β-GD and endophytic fungi leads to higher fungal colonization. Overall, this article indicates fungi have evolved to utilize a plant defense-related enzyme to subvert plant immunity. Summary by Kamal Kumar Malukani, @KamalMalukani. Plant Cell 10.1093/plcell/koac114
The studies on the apoplastic interactions of plants and fungi often focus on the fungal cell-wall, but recent studies indicate various plant colonizing fungi also have an exopolysaccharide (EPS) layer outside their cell wall. Chandrasekar et al. observed that the composition of this fungal EPS is distinct from the fungal cell wall and the secreted culture filtrate, indicating it is neither just an extension of the cell wall nor something simply secreted out into the medium. The activity of a barley glucanase (which is secreted in response to fungal treatment) on the EPS that is secreted by endophytic or pathogenic fungi leads to the release of a β-glucan decasaccharide (β-GD). Interestingly application of β-GD to barley or Arabidopsis suppresses the immune-responsive ROS burst triggered by other elicitors of immunity, but it does not affect other early immune responses. In vitro studies indicate β-GD can scavenge ROS indicating it might be quenching apoplastic ROS that is secreted by plants during plant-microbe interactions. Co-treatment of plants with β-GD and endophytic fungi leads to higher fungal colonization. Overall, this article indicates fungi have evolved to utilize a plant defense-related enzyme to subvert plant immunity. Summary by Kamal Kumar Malukani, @KamalMalukani. Plant Cell 10.1093/plcell/koac114