Plant Science Research Weekly: May 10th
Durum wheat genome highlights past domestication signatures and future improvement targets
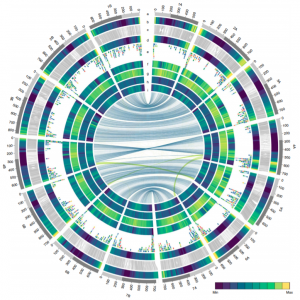 Durum wheat cultivar Svevo (Triticum turgidum L. ssp. durum) is a cereal crop predominantly grown for pasta production. It is the result of multiple rounds of domestication, originally deriving from wild emmer wheat from the Fertile Crescent approximately 10,000 years ago. Here, Maccaferri et al. report the high quality genome of durum wheat, creating an indispensable resource to study wheat evolution and domestication. The use of comparative analysis to assess the impact of thousands of years of selective breeding reveals genome wide modifications as well as QTLs for agriculturally important traits. This genome alongside genotypic, gene mapping and expression data will be a valuable tool for the wheat and crop community e.g., gene discovery for grain quality and quantity (Summary by Alex Bowles) Nature Genetics 10.1038/s41588-019-0381-3.
Durum wheat cultivar Svevo (Triticum turgidum L. ssp. durum) is a cereal crop predominantly grown for pasta production. It is the result of multiple rounds of domestication, originally deriving from wild emmer wheat from the Fertile Crescent approximately 10,000 years ago. Here, Maccaferri et al. report the high quality genome of durum wheat, creating an indispensable resource to study wheat evolution and domestication. The use of comparative analysis to assess the impact of thousands of years of selective breeding reveals genome wide modifications as well as QTLs for agriculturally important traits. This genome alongside genotypic, gene mapping and expression data will be a valuable tool for the wheat and crop community e.g., gene discovery for grain quality and quantity (Summary by Alex Bowles) Nature Genetics 10.1038/s41588-019-0381-3.
Structural and functional imaging of large and opaque plant specimen
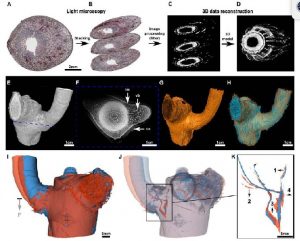 Even the simplest organisms are highly complex systems in which countless dynamic biochemical processes occur simultaneously. To fully understand the mechanical significance of such a complex molecular machine, the ability to accurately characterize dynamic processes at different scales is required. A variety of imaging methodologies are used to collect data for quantitative studies of plant growth and development. Multi-level data can be acquired, from macroscopic (m to cm) to molecular (nm), and from weeks to seconds. Static (at a given time; 3D) and dynamic (at different time points; quasi 4D) structural and functional imaging are a prerequisite for spatially resolving the form-structure-function relationships in plants. Each imaging technique has specific advantages and limitations as well as the appropriate fields of application. In this article, Hesse et al. described three different methods (LMTS, µ-CT and MRI) used in plant sciences, making a comparison of the imaging principles with a particular focus on their use in imaging large and opaque plant specimens. (Summarized by Francesca Resentini) J. Exp. Bot. 10.1093/jxb/erz186
Even the simplest organisms are highly complex systems in which countless dynamic biochemical processes occur simultaneously. To fully understand the mechanical significance of such a complex molecular machine, the ability to accurately characterize dynamic processes at different scales is required. A variety of imaging methodologies are used to collect data for quantitative studies of plant growth and development. Multi-level data can be acquired, from macroscopic (m to cm) to molecular (nm), and from weeks to seconds. Static (at a given time; 3D) and dynamic (at different time points; quasi 4D) structural and functional imaging are a prerequisite for spatially resolving the form-structure-function relationships in plants. Each imaging technique has specific advantages and limitations as well as the appropriate fields of application. In this article, Hesse et al. described three different methods (LMTS, µ-CT and MRI) used in plant sciences, making a comparison of the imaging principles with a particular focus on their use in imaging large and opaque plant specimens. (Summarized by Francesca Resentini) J. Exp. Bot. 10.1093/jxb/erz186
Shared expression of crassulacean acid metabolism (CAM) genes pre-dates the origin of CAM in the genus Yucca
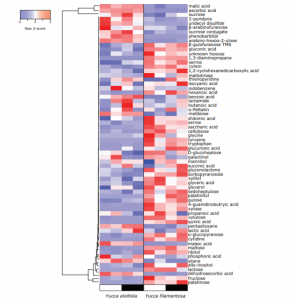 Crassulacean acid metabolism (CAM) is a carbon fixation pathway that reduces photorespiration and increases water use efficiency, enabling CAM plants to survive in inhospitable environments. The evolution of CAM on 35 independent occasions across angiosperms makes it a notable case study of convergent evolution. Here, Heyduk et al. perform transcriptomic and metabolomic analysis on a time course dataset of three Yucca (Asparagaceae) species, representing C3, CAM and C3/CAM intermediate photosynthetic strategies. Distinct transcript profiles revealed differences in gene expression of certain core CAM genes between C3 and CAM species. Metabolomic variation identified differences of starch and soluble sugar metabolism, highlighting the fitness costs associated with CAM. However, similarities of particular CAM genes (e.g. PEPC) in C3 and CAM plants at the expression level were identified suggesting the presence of shared traits in the ancestral Yucca genome which enabled the evolution of CAM photosynthesis. (Summary by Alex Bowles) J. Exp. Bot. 10.1093/jxb/erz105
Crassulacean acid metabolism (CAM) is a carbon fixation pathway that reduces photorespiration and increases water use efficiency, enabling CAM plants to survive in inhospitable environments. The evolution of CAM on 35 independent occasions across angiosperms makes it a notable case study of convergent evolution. Here, Heyduk et al. perform transcriptomic and metabolomic analysis on a time course dataset of three Yucca (Asparagaceae) species, representing C3, CAM and C3/CAM intermediate photosynthetic strategies. Distinct transcript profiles revealed differences in gene expression of certain core CAM genes between C3 and CAM species. Metabolomic variation identified differences of starch and soluble sugar metabolism, highlighting the fitness costs associated with CAM. However, similarities of particular CAM genes (e.g. PEPC) in C3 and CAM plants at the expression level were identified suggesting the presence of shared traits in the ancestral Yucca genome which enabled the evolution of CAM photosynthesis. (Summary by Alex Bowles) J. Exp. Bot. 10.1093/jxb/erz105
Land plants recruited an ancestral bHLH for tip-growing surface cell development ($)
 Land plants (embryophytes) evolved from freshwater charophycean algae over 450 million years ago. The transition from aquatic to terrestrial environments likely required the evolution and expansion of genetic programs controlling three dimensional growth and the formation of tip-growing surface cells (rhizoids/root hairs) from the plant body. Bonnot et al. investigated the function and evolutionary history of basic helix-loop-helix (bHLH) transcription factors implicated in the development of tip-growing surface cells in land plants and their algal predecessors. Phylogenetic analyses first established that the multicellular charophytes Chara braunii and Coleochate nitellarum encode homologs belonging to the bHLH VIII gene family that contains the tip-growth (root hair) controlling members RHD6 (ROOT HAIR DEFECTIVE6) and RSL (RHD6-LIKE). Cross-complementation experiments utilizing the root hair defective Arabidopsis thaliana (angiosperm) double mutant rhd6/rsl1 and the rhizoid-deficient Marchantia polymorpha (liverwort) Mprsl1 mutant showed that ectopic expression of charophyte bHLHVIII homologs failed to rescue the root hair/rhizoid-defective phenotypes of mutants in either lineage, whereas MpRSL expression reliably rescued Arabidopsis rhd6/rsl1 and Mprsl. To identify potential roles for charophyte bHLHVIII genes in development, the authors performed expression analyses in distinct tissues of the model charophyte Chara braunii. Intriguingly, CbbHLHVIII was minimally expressed in C. braunii rhizoids and was most highly abundant in gametangia (sexual reproduction) and thalli (body). More detailed analysis further revealed the preferential expression of CbbHLHVIII in young branchletes with gametangia and younger tissues of the thallus (newly developed nodes) containing apical cells. Collectively, these data clarify the evolutionary history of bHLHVIII transcription factors that underwent neofunctionalization in land plants to control the development of rhizoids and root hairs. (Summary by Phil Carella) New Phytol. 10.1111/nph.15829
Land plants (embryophytes) evolved from freshwater charophycean algae over 450 million years ago. The transition from aquatic to terrestrial environments likely required the evolution and expansion of genetic programs controlling three dimensional growth and the formation of tip-growing surface cells (rhizoids/root hairs) from the plant body. Bonnot et al. investigated the function and evolutionary history of basic helix-loop-helix (bHLH) transcription factors implicated in the development of tip-growing surface cells in land plants and their algal predecessors. Phylogenetic analyses first established that the multicellular charophytes Chara braunii and Coleochate nitellarum encode homologs belonging to the bHLH VIII gene family that contains the tip-growth (root hair) controlling members RHD6 (ROOT HAIR DEFECTIVE6) and RSL (RHD6-LIKE). Cross-complementation experiments utilizing the root hair defective Arabidopsis thaliana (angiosperm) double mutant rhd6/rsl1 and the rhizoid-deficient Marchantia polymorpha (liverwort) Mprsl1 mutant showed that ectopic expression of charophyte bHLHVIII homologs failed to rescue the root hair/rhizoid-defective phenotypes of mutants in either lineage, whereas MpRSL expression reliably rescued Arabidopsis rhd6/rsl1 and Mprsl. To identify potential roles for charophyte bHLHVIII genes in development, the authors performed expression analyses in distinct tissues of the model charophyte Chara braunii. Intriguingly, CbbHLHVIII was minimally expressed in C. braunii rhizoids and was most highly abundant in gametangia (sexual reproduction) and thalli (body). More detailed analysis further revealed the preferential expression of CbbHLHVIII in young branchletes with gametangia and younger tissues of the thallus (newly developed nodes) containing apical cells. Collectively, these data clarify the evolutionary history of bHLHVIII transcription factors that underwent neofunctionalization in land plants to control the development of rhizoids and root hairs. (Summary by Phil Carella) New Phytol. 10.1111/nph.15829
Post-transcriptional regulation of FLOWERING LOCUS T modulates heat-dependent source-sink development in potato
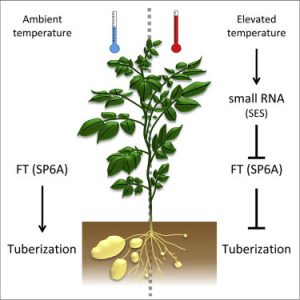 Tuber formation (tuberization) in potatoes is controlled by both endogenous and environmental signals particularly day length and temperature. Long days and high temperature impede tuberization while it is enhanced by short days and low temperature. When conditions are favourable for tuberization, shoot-derived mobile signals are translocated into the root where tuber formation is activated. It was previously shown that the FLOWERING LOCUS T (FT) homolog SP6A encodes one of these tuberization signals. In addition, SP6A expression is controlled by both day-length and temperature. However, the molecular mechanism mediating this outcome is unknown. Lehretz et al. showed that the overexpression of a codon optimized SP6A resulted in early tuberization and additional tuber growth as a result of altered meristem identity in potato. They identified a novel small RNA that targets SP6A. Its expression increases under elevated temperature and downregulates the expression of SP6A. By overexpressing a target mimicry construct, higher transcript level of SP6A and tuberization was observed even at elevated temperature, suggesting that inhibition of tuberization at high temperature as a result of downregulated SP6A is modulated by a small RNA. Although the detailed mechanism of this small RNA action is not yet known, the authors elegantly show that SP6A is regulated at the post-transcriptional level by a temperature controlled small RNA. (Summary by Toluwase Olukayode). Curr. Biol. 10.1016/j.cub.2019.04.027
Tuber formation (tuberization) in potatoes is controlled by both endogenous and environmental signals particularly day length and temperature. Long days and high temperature impede tuberization while it is enhanced by short days and low temperature. When conditions are favourable for tuberization, shoot-derived mobile signals are translocated into the root where tuber formation is activated. It was previously shown that the FLOWERING LOCUS T (FT) homolog SP6A encodes one of these tuberization signals. In addition, SP6A expression is controlled by both day-length and temperature. However, the molecular mechanism mediating this outcome is unknown. Lehretz et al. showed that the overexpression of a codon optimized SP6A resulted in early tuberization and additional tuber growth as a result of altered meristem identity in potato. They identified a novel small RNA that targets SP6A. Its expression increases under elevated temperature and downregulates the expression of SP6A. By overexpressing a target mimicry construct, higher transcript level of SP6A and tuberization was observed even at elevated temperature, suggesting that inhibition of tuberization at high temperature as a result of downregulated SP6A is modulated by a small RNA. Although the detailed mechanism of this small RNA action is not yet known, the authors elegantly show that SP6A is regulated at the post-transcriptional level by a temperature controlled small RNA. (Summary by Toluwase Olukayode). Curr. Biol. 10.1016/j.cub.2019.04.027
Calcium-promoted interaction between the C2-domain protein EHB1 and metal transporter IRT1 inhibits Arabidopsis iron acquisition
 Iron uptake is tightly controlled so that the plant takes up not too much but not too little. Khan et al. used a yeast two-hybrid screen to search for proteins that interact with the iron transporter IRT1. They identified a protein previously shown to be involved in the hypocotyl bending response, EHB1 (ENHANCED BENDING1), which has a calcium and lipid binding domain. Their results suggest that EHB1 and IRT1 interact in a calcium-dependent manner at the plasma membrane. The authors also performed molecular dynamics simulations of EHB1 binding to the membrane, which suggest that it interacts via the calcium-binding site. Under low iron conditions, loss-of-function ehb1 mutants appear less iron-deficient, suggesting that EHB1 is a negative regulator of iron uptake. The authors demonstrate that through its binding, EHB1 directly inhibits IRT1 function. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00163
Iron uptake is tightly controlled so that the plant takes up not too much but not too little. Khan et al. used a yeast two-hybrid screen to search for proteins that interact with the iron transporter IRT1. They identified a protein previously shown to be involved in the hypocotyl bending response, EHB1 (ENHANCED BENDING1), which has a calcium and lipid binding domain. Their results suggest that EHB1 and IRT1 interact in a calcium-dependent manner at the plasma membrane. The authors also performed molecular dynamics simulations of EHB1 binding to the membrane, which suggest that it interacts via the calcium-binding site. Under low iron conditions, loss-of-function ehb1 mutants appear less iron-deficient, suggesting that EHB1 is a negative regulator of iron uptake. The authors demonstrate that through its binding, EHB1 directly inhibits IRT1 function. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00163
NRT1.1B is associated with root microbiota composition and nitrogen use in field grown rice ($)
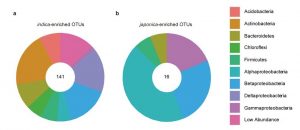 Availability of nitrogen has always been a limiting factor for rice production. Different soil inhabitants and root-associated bacterial populations are involved in making nitrogen available to plants in organic as well as in inorganic forms. However, plant absorption of nitrogen is selective and affects its nitrogen use efficiency (NUE). The two main Asian cultivated rice varieties, japonica and indica have different NUE, attributed in part to the natural variation in a gene called NRT1.1B that encodes a nitrate transporter and sensor. By using 68 indica and 27 japonica varieties, Zhang et al. found a relationship between NRT1.1B and rice root-associated microbial populations. The microbiome community structure analysis from both varieties showed a clear influence of genotype effect, moreover the indica variety showed higher microbial diversity than japonica. Functional assignment to the microbial population showed more of the indica associated microbes are involved in nitrogen metabolism function. The authors also generated a synthetic microbial community based on genotype-specific and NRT1.1B-associated observations by using established culture collections. (Summary by Mugdha Sabale) Nature Biotechnol. 10.1038/s41587-019-0104-4
Availability of nitrogen has always been a limiting factor for rice production. Different soil inhabitants and root-associated bacterial populations are involved in making nitrogen available to plants in organic as well as in inorganic forms. However, plant absorption of nitrogen is selective and affects its nitrogen use efficiency (NUE). The two main Asian cultivated rice varieties, japonica and indica have different NUE, attributed in part to the natural variation in a gene called NRT1.1B that encodes a nitrate transporter and sensor. By using 68 indica and 27 japonica varieties, Zhang et al. found a relationship between NRT1.1B and rice root-associated microbial populations. The microbiome community structure analysis from both varieties showed a clear influence of genotype effect, moreover the indica variety showed higher microbial diversity than japonica. Functional assignment to the microbial population showed more of the indica associated microbes are involved in nitrogen metabolism function. The authors also generated a synthetic microbial community based on genotype-specific and NRT1.1B-associated observations by using established culture collections. (Summary by Mugdha Sabale) Nature Biotechnol. 10.1038/s41587-019-0104-4
Slow canopy wilting enhances drought-tolerance in soybean
 As plants are sessile organisms, they have developed highly sophisticated mechanisms to allow the modulation of development in response to environmental changes, thus maximizing their chance of survival. When soil dries, soybeans with a slow canopy wilting (SW) phenotype have delayed canopy/leaf wilting to overcome the negative impacts of drought stress. Two exotic (landraces) plant introductions (PI 567690 and PI 567731) were identified as new SW lines. Heng et al. studied the physiological mechanism of these two SW lines, trying to explain the effects of SW on yield of soybean under stress. They found that the SW phenotype in these exotic lines is involved in water use efficiency and conservation, as a result of limited transpiration response induced by water deficiency in soil. Moreover, the mechanism behind this may not be the previously hypothesized lack of silver-sensitive aquaporins, but more likely may be linked with a larger and deeper root system. Genetic analysis identified new QTLs associated with the slow canopy wilting phenotype. This study also confirmed the importance of SW on yield protection under drought stress. (Summary by Nanxun Qin). J. Exp. Bot. 10.1093/jxb/erz150
As plants are sessile organisms, they have developed highly sophisticated mechanisms to allow the modulation of development in response to environmental changes, thus maximizing their chance of survival. When soil dries, soybeans with a slow canopy wilting (SW) phenotype have delayed canopy/leaf wilting to overcome the negative impacts of drought stress. Two exotic (landraces) plant introductions (PI 567690 and PI 567731) were identified as new SW lines. Heng et al. studied the physiological mechanism of these two SW lines, trying to explain the effects of SW on yield of soybean under stress. They found that the SW phenotype in these exotic lines is involved in water use efficiency and conservation, as a result of limited transpiration response induced by water deficiency in soil. Moreover, the mechanism behind this may not be the previously hypothesized lack of silver-sensitive aquaporins, but more likely may be linked with a larger and deeper root system. Genetic analysis identified new QTLs associated with the slow canopy wilting phenotype. This study also confirmed the importance of SW on yield protection under drought stress. (Summary by Nanxun Qin). J. Exp. Bot. 10.1093/jxb/erz150
A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls
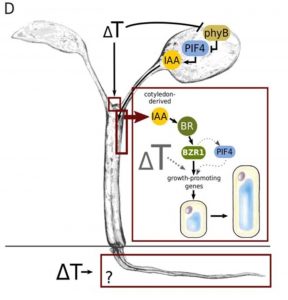 An increase in ambient temperature affects plant growth and development; for instance, high temperatures trigger elongation in petioles, hypocotyls, and roots in Arabidopsis seedlings. Bellstaedt et al. studied the spatial relationships in sensing, signaling and growth responses to high temperature in these tissues. The first clue about organ-specific processes came from the transcriptomic analysis when they identified differential expression of hormone-response genes in cotyledon vs root samples. The authors focused on auxin as a signal, since previous reports indicate PIF4 (PHYTOCHROME-INTERACTING FACTOR 4, a thermomorphogenesis-signaling hub), induces auxin biosynthesis genes, and mutants affected in the production (yuc1-D) or transport (pin) of auxins showed altered response to hypocotyl elongation in high temperatures. They dissected tissues and analyzed the individual response of hypocotyls and roots. Detached roots elongated in response to high temperatures, but hypocotyls did not show the thermomorphogenic response. The crucial role of auxin was further confirmed with the application of an auxin transporter inhibitor (NPA), which caused the same effect of the cotyledon detachment. Additional players in this process involve components of the brassinosteroid signaling pathway. The authors conclude that cotyledons sense high temperatures and generate an auxin signal that is translocated to the hypocotyls where it triggers cell elongation. (Summary by Humberto Herrera-Ubaldo) Plant Phys. 10.1104/pp.18.01377
An increase in ambient temperature affects plant growth and development; for instance, high temperatures trigger elongation in petioles, hypocotyls, and roots in Arabidopsis seedlings. Bellstaedt et al. studied the spatial relationships in sensing, signaling and growth responses to high temperature in these tissues. The first clue about organ-specific processes came from the transcriptomic analysis when they identified differential expression of hormone-response genes in cotyledon vs root samples. The authors focused on auxin as a signal, since previous reports indicate PIF4 (PHYTOCHROME-INTERACTING FACTOR 4, a thermomorphogenesis-signaling hub), induces auxin biosynthesis genes, and mutants affected in the production (yuc1-D) or transport (pin) of auxins showed altered response to hypocotyl elongation in high temperatures. They dissected tissues and analyzed the individual response of hypocotyls and roots. Detached roots elongated in response to high temperatures, but hypocotyls did not show the thermomorphogenic response. The crucial role of auxin was further confirmed with the application of an auxin transporter inhibitor (NPA), which caused the same effect of the cotyledon detachment. Additional players in this process involve components of the brassinosteroid signaling pathway. The authors conclude that cotyledons sense high temperatures and generate an auxin signal that is translocated to the hypocotyls where it triggers cell elongation. (Summary by Humberto Herrera-Ubaldo) Plant Phys. 10.1104/pp.18.01377
Thermal response in plants: leaf hyponasty
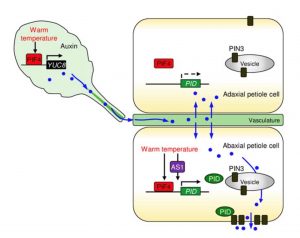 Non-directional stimuli can trigger directional movements in plant organs. For instance, high temperature causes the upward bending of leaf petioles, a process known as leaf hyponasty, which helps to cool the leaves. In this study Park et al. explored the link between leaf thermonasty and auxins. Gene expression analysis under thermal induction revealed that SAUR gene expression is higher in leaf petiole abaxial samples compared to the adaxial ones. Using the DR5:GUS auxin reporter line, the authors found a preferential auxin accumulation in the abaxial petiole region, suggesting that auxin is involved in the thermal regulation of leaf hyponasty. Other players are involved in the process including ASYMMETRIC LEAVES 1 (AS1, a master regulator of leaf polarity developmental programs), and additional auxin signaling components, for instance, PID (the kinase that regulates the PIN3 transporter). In their model, PIF4-triggers auxin production in leaf blade and its transport to the petiole, AS1 directs PID transcription in the abaxial petiole, and PID-mediated PIN3 polarization determines the direction of leaf bending. (Summary by Humberto Herrera-Ubaldo) Plant Physiol. 10.1104/pp.19.00139
Non-directional stimuli can trigger directional movements in plant organs. For instance, high temperature causes the upward bending of leaf petioles, a process known as leaf hyponasty, which helps to cool the leaves. In this study Park et al. explored the link between leaf thermonasty and auxins. Gene expression analysis under thermal induction revealed that SAUR gene expression is higher in leaf petiole abaxial samples compared to the adaxial ones. Using the DR5:GUS auxin reporter line, the authors found a preferential auxin accumulation in the abaxial petiole region, suggesting that auxin is involved in the thermal regulation of leaf hyponasty. Other players are involved in the process including ASYMMETRIC LEAVES 1 (AS1, a master regulator of leaf polarity developmental programs), and additional auxin signaling components, for instance, PID (the kinase that regulates the PIN3 transporter). In their model, PIF4-triggers auxin production in leaf blade and its transport to the petiole, AS1 directs PID transcription in the abaxial petiole, and PID-mediated PIN3 polarization determines the direction of leaf bending. (Summary by Humberto Herrera-Ubaldo) Plant Physiol. 10.1104/pp.19.00139
Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses
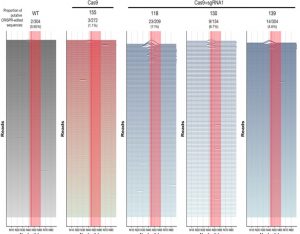 CRISPR/Cas9 is a promising gene editing tool that has already been successfully used to modify many plant genes. In these applications, the gene editing machinery is transiently employed to make a stable genomic change which is then passed on to the progeny. A different application is to use CRISPR/Cas9 for defense against viruses; after all, this tool is derived from a bacterial viral defense system. In this latter application, the CRISPR/Cas9 system is continually expressed and available to identify and cleave viral DNA. Previous studies have suggested that CRISPR/Cas9 is effective in this way, but a new study by Mehta et al. casts doubt. They show that when cassava is engineered to employ CRISPR/Cas to resist cassava mosaic virus the viruses rapidly mutate to prevent cleavage; the viruses become resistant to the defense machinery. The authors conclude, “the implementation of technologies with the potential to speed up virus evolution should be carefully assessed as they pose significant biosafety risks.”(Summary by Mary Williams) Genome Biol. 10.1186/s13059-019-1678-3
CRISPR/Cas9 is a promising gene editing tool that has already been successfully used to modify many plant genes. In these applications, the gene editing machinery is transiently employed to make a stable genomic change which is then passed on to the progeny. A different application is to use CRISPR/Cas9 for defense against viruses; after all, this tool is derived from a bacterial viral defense system. In this latter application, the CRISPR/Cas9 system is continually expressed and available to identify and cleave viral DNA. Previous studies have suggested that CRISPR/Cas9 is effective in this way, but a new study by Mehta et al. casts doubt. They show that when cassava is engineered to employ CRISPR/Cas to resist cassava mosaic virus the viruses rapidly mutate to prevent cleavage; the viruses become resistant to the defense machinery. The authors conclude, “the implementation of technologies with the potential to speed up virus evolution should be carefully assessed as they pose significant biosafety risks.”(Summary by Mary Williams) Genome Biol. 10.1186/s13059-019-1678-3



