Plant Science Research Weekly: March 18, 2022
Review: A rulebook for peptide control of legume–microbe endosymbiosis
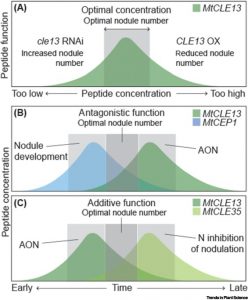 Symbiotic associations with bacterial or fungal partners enhance nutrient uptake for most plants, and recent years have uncovered the very sophisticated means by which these associations are established and controlled. Peptides have emerged as key regulators of many facets of mycorrhizal and rhizobial symbioses. Here, Roy and Müller synthesize much of our current understanding of these processes and identify five key “rules” to describe them. 1) Multiple peptides work in concert to fine-tune processes; 2) Some of these converge on common downstream hubs; 3) Peptide signals intersect with the classic hormones; 4) Symbiotic-associated peptides often are found in large gene families; 5) (not surprisingly) Microbes synthesize peptide mimics to enhance colonization. This is a thoughtful, comprehensive review that provides an important framework for understanding these crucial interactions. (Summary by Mary Williams @PlantTeaching). Trends Plant Sci. 10.1016/j.tplants.2022.02.002
Symbiotic associations with bacterial or fungal partners enhance nutrient uptake for most plants, and recent years have uncovered the very sophisticated means by which these associations are established and controlled. Peptides have emerged as key regulators of many facets of mycorrhizal and rhizobial symbioses. Here, Roy and Müller synthesize much of our current understanding of these processes and identify five key “rules” to describe them. 1) Multiple peptides work in concert to fine-tune processes; 2) Some of these converge on common downstream hubs; 3) Peptide signals intersect with the classic hormones; 4) Symbiotic-associated peptides often are found in large gene families; 5) (not surprisingly) Microbes synthesize peptide mimics to enhance colonization. This is a thoughtful, comprehensive review that provides an important framework for understanding these crucial interactions. (Summary by Mary Williams @PlantTeaching). Trends Plant Sci. 10.1016/j.tplants.2022.02.002
A dominant mutation in β-AMYLASE1 disrupts nighttime control of starch degradation in Arabidopsis leaves
 Plants use starch in leaf chloroplasts to fuel their growth and metabolism during the night. In Arabidopsis, nighttime starch degradation is tightly regulated; the degradation rate is linear and adjusts to both the availability of starch and length of night, so that most but not all the starch is used up by dawn. The rate, S/T, is determined by a hypothetical “S” which tracks the amount of starch remaining, and “T”, the time till dawn sensed by the plant. Here, Feike et al. identified the bam1-2D mutant of BETA-AMYLASE1 (BAM1), which has a dominant missense mutation (S132N) causing accelerated starch degradation and premature exhaustion of starch reserves, possibly due to an uncoupling of S from the actual starch content. Mutant studies show that BAM1 transcript levels, protein content or phosphorylation state are not the cause, and in vitro enzymatic activity is not affected either. Instead, the strong phenotype could be caused by conformational changes leading to altered protein-protein interactions of a complex in vivo. Previous studies showed that BAM1 associates with a putative scaffold protein, LIKE SEX FOUR1 (LSF1), and that lsf1 mutants have a starch excess phenotype. Here, native gels, immunoblots and starch degradation assays from leaf extracts of various starch degradation mutants show an enhanced association of BAM1(S132N) with LSF1 compared to wild-type BAM1, and that this association is necessary for the accelerated starch degradation phenotype. Interestingly, bam1-2D lsf1 double mutants restore starch degradation to wild type rates, indicating that the bam1-2D mutation also overcomes the need for LSF1 in regular starch degradation. (Summary by Jiawen Chen @Jiaaawen) Plant Physiol. 10.1093/plphys/kiab603
Plants use starch in leaf chloroplasts to fuel their growth and metabolism during the night. In Arabidopsis, nighttime starch degradation is tightly regulated; the degradation rate is linear and adjusts to both the availability of starch and length of night, so that most but not all the starch is used up by dawn. The rate, S/T, is determined by a hypothetical “S” which tracks the amount of starch remaining, and “T”, the time till dawn sensed by the plant. Here, Feike et al. identified the bam1-2D mutant of BETA-AMYLASE1 (BAM1), which has a dominant missense mutation (S132N) causing accelerated starch degradation and premature exhaustion of starch reserves, possibly due to an uncoupling of S from the actual starch content. Mutant studies show that BAM1 transcript levels, protein content or phosphorylation state are not the cause, and in vitro enzymatic activity is not affected either. Instead, the strong phenotype could be caused by conformational changes leading to altered protein-protein interactions of a complex in vivo. Previous studies showed that BAM1 associates with a putative scaffold protein, LIKE SEX FOUR1 (LSF1), and that lsf1 mutants have a starch excess phenotype. Here, native gels, immunoblots and starch degradation assays from leaf extracts of various starch degradation mutants show an enhanced association of BAM1(S132N) with LSF1 compared to wild-type BAM1, and that this association is necessary for the accelerated starch degradation phenotype. Interestingly, bam1-2D lsf1 double mutants restore starch degradation to wild type rates, indicating that the bam1-2D mutation also overcomes the need for LSF1 in regular starch degradation. (Summary by Jiawen Chen @Jiaaawen) Plant Physiol. 10.1093/plphys/kiab603
A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants
 Single-cell RNA sequencing (scRNA-seq) is a powerful tool to identify rare and novel cell types, characterize multiple cell types, and more accurately understand their functions across their life cycles. The Arabidopsis root is a perfect system in which to investigate cellular differentiation due to its immobile and organized cells. Shahan and Hsu et al. built a root scRNA-seq atlas (>110 000 cells) using an optimal transport-base method, StationaryOT. This method allows the reconstruction of developmental trajectories and helps to understand how the fate acquisition of a specific cell type is related to other cell types. The authors tested the utility of this method by performing scRNA-seq of shortroot (shr) and scarecrow (scr) mutants to identify unknown developmental phenotypes. scRNA-seq and in vivo experiments of scr suggest that young cells are cortex-like while older cells changes their identity to endodermis-like. These findings show how cell identity acquires attributes of other cell types over time. This work provides an outlook to test unknown cell-type specific markers and identify cell-type-specific responses to different environmental conditions. (Summary by Andrea Gómez Felipe @andreagomezfe) Dev. Cell 10.1016/j.devcel.2022.01.008
Single-cell RNA sequencing (scRNA-seq) is a powerful tool to identify rare and novel cell types, characterize multiple cell types, and more accurately understand their functions across their life cycles. The Arabidopsis root is a perfect system in which to investigate cellular differentiation due to its immobile and organized cells. Shahan and Hsu et al. built a root scRNA-seq atlas (>110 000 cells) using an optimal transport-base method, StationaryOT. This method allows the reconstruction of developmental trajectories and helps to understand how the fate acquisition of a specific cell type is related to other cell types. The authors tested the utility of this method by performing scRNA-seq of shortroot (shr) and scarecrow (scr) mutants to identify unknown developmental phenotypes. scRNA-seq and in vivo experiments of scr suggest that young cells are cortex-like while older cells changes their identity to endodermis-like. These findings show how cell identity acquires attributes of other cell types over time. This work provides an outlook to test unknown cell-type specific markers and identify cell-type-specific responses to different environmental conditions. (Summary by Andrea Gómez Felipe @andreagomezfe) Dev. Cell 10.1016/j.devcel.2022.01.008
The miRNome function transitions from regulating developmental genes to transposable elements during pollen maturation
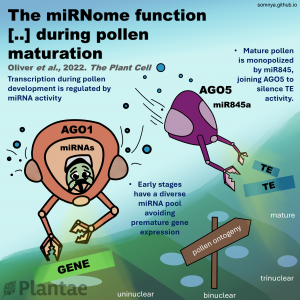 Genes (protein-coding or noncoding) and transposable elements (TEs) are the main characters in the genomic landscape, but there are few reported systems where we have seen all acting together to produce a phenotype. Oliver and collaborators analyzed miRNA populations during pollen maturation in an outstanding experimental and computational initiative. In Arabidopsis, pollen development goes from uninuclear to binuclear, trinuclear, and finally, mature pollen. Initially, the pool of miRNAs is diverse at the uninuclear stage but it becomes dominated by miR845 at the mature stage. Different miRNAs are selectively introduced to Argonaute (AGO) proteins depending on their structure and which target they pursue. AGO1 is commonly present in the uninuclear stage, repressing coding transcripts related to molecular functions. As pollen matures, AGO5 appears and associates with miR845a to cleave TEs. Jointly, miRNAs and AGO proteins build a bivalent system that starts in gene regulation and ends in TE post-transcriptional silencing and, presumably, siRNA production. Many exciting questions arise from this work, such as is it a conserved ontogenetic system in plants? Could plant development be predicted by looking at the genomic and transcriptomic landscape? This unique work should be introduced into each plant physiology and genomics course. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) Plant Cell 10.1093/plcell/koab280
Genes (protein-coding or noncoding) and transposable elements (TEs) are the main characters in the genomic landscape, but there are few reported systems where we have seen all acting together to produce a phenotype. Oliver and collaborators analyzed miRNA populations during pollen maturation in an outstanding experimental and computational initiative. In Arabidopsis, pollen development goes from uninuclear to binuclear, trinuclear, and finally, mature pollen. Initially, the pool of miRNAs is diverse at the uninuclear stage but it becomes dominated by miR845 at the mature stage. Different miRNAs are selectively introduced to Argonaute (AGO) proteins depending on their structure and which target they pursue. AGO1 is commonly present in the uninuclear stage, repressing coding transcripts related to molecular functions. As pollen matures, AGO5 appears and associates with miR845a to cleave TEs. Jointly, miRNAs and AGO proteins build a bivalent system that starts in gene regulation and ends in TE post-transcriptional silencing and, presumably, siRNA production. Many exciting questions arise from this work, such as is it a conserved ontogenetic system in plants? Could plant development be predicted by looking at the genomic and transcriptomic landscape? This unique work should be introduced into each plant physiology and genomics course. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) Plant Cell 10.1093/plcell/koab280
The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination
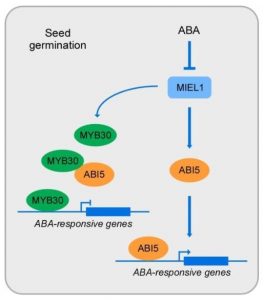 Abscisic acid (ABA) is a phytohormone that plays a crucial role in the inhibition of seed germination, and the transcription factor ABI5 is the final and common downstream factor that represses germination. Many proteins are known to inhibit ABI5, such as the mediator complex subunit MED25 or other transcription factors, but how these proteins interact and function together to properly regulate ABA response levels during germination is still unknown. In this work, Nie et al. suggest a regulatory module that precisely regulates ABA signaling during seed germination by controlling ABI5 activity. The authors identified a RING-type ubiquitin E3 ligase MYB30-INTERACTING E3 LIGASE 1 (MIEL1) and its target MYB30 as ABI5-interacting proteins. For seed germination to proceed, the inhibition of MIEL1 releases MYB30, which restricts ABA responses by inhibiting both the activity of ABI5 and the transcription of ABA-responsive genes. If ABA accumulates, MIEL1 shows decreased stability, which also produces a weakened interaction between MIEL1 and ABI5, releasing ABI5 and the transcription of ABA-responsive genes. This work sheds light on the stability and transcriptional activity of ABI5 during germination. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) J. Integr. Plant Biol. 10.1111/jipb.13234
Abscisic acid (ABA) is a phytohormone that plays a crucial role in the inhibition of seed germination, and the transcription factor ABI5 is the final and common downstream factor that represses germination. Many proteins are known to inhibit ABI5, such as the mediator complex subunit MED25 or other transcription factors, but how these proteins interact and function together to properly regulate ABA response levels during germination is still unknown. In this work, Nie et al. suggest a regulatory module that precisely regulates ABA signaling during seed germination by controlling ABI5 activity. The authors identified a RING-type ubiquitin E3 ligase MYB30-INTERACTING E3 LIGASE 1 (MIEL1) and its target MYB30 as ABI5-interacting proteins. For seed germination to proceed, the inhibition of MIEL1 releases MYB30, which restricts ABA responses by inhibiting both the activity of ABI5 and the transcription of ABA-responsive genes. If ABA accumulates, MIEL1 shows decreased stability, which also produces a weakened interaction between MIEL1 and ABI5, releasing ABI5 and the transcription of ABA-responsive genes. This work sheds light on the stability and transcriptional activity of ABI5 during germination. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) J. Integr. Plant Biol. 10.1111/jipb.13234
Nucleocytoplasmic shuttling of ETHYLENE RESPONSE FACTOR 5 mediated by nitric oxide suppresses ethylene biosynthesis in apple fruit
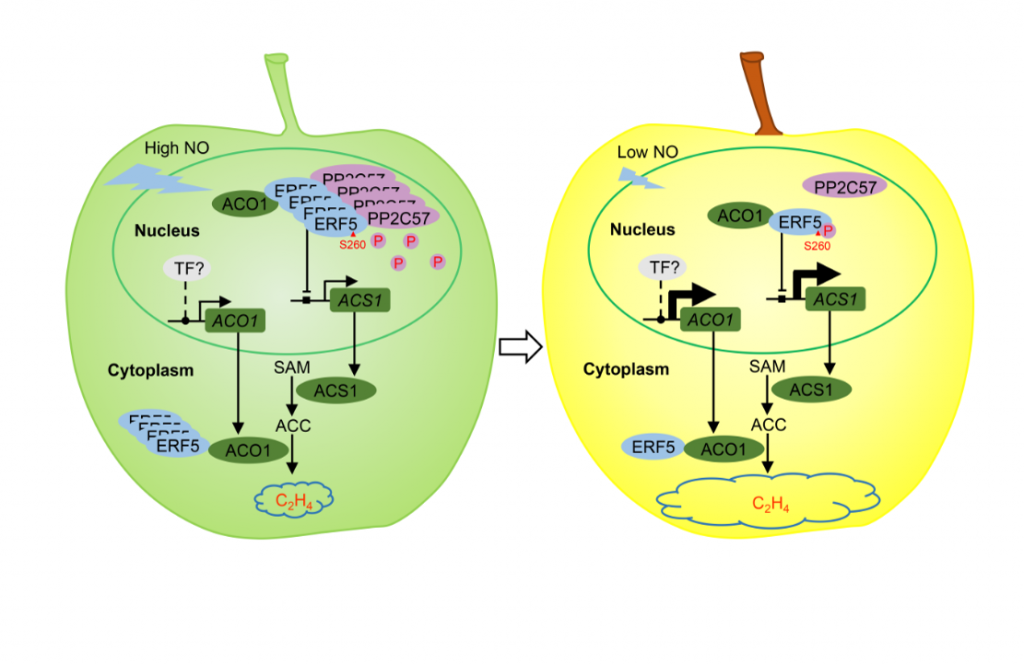 Mechanisms of phytohormone modulation and their interface with other regulatory molecules are still being uncovered. Nitric oxide (NO) is a regulatory molecule which has been shown to affect ethylene biosynthesis through the action of ETHYLENE RESPONSE FACTOR 5 (ERF5) in apple fruit. New work by Ji et al. demonstrates that ERF5 nucleocytoplasmic shuttling is dependent on its phosphorylation state. Following application of a NO donor, ERF5 expression increases while ethylene biosynthesis is inhibited. NO stimulates PP2C57-mediated dephosphorylation of ERF5 and facilitates ERF5 translocation from the nucleus to cytoplasm where it suppresses ethylene biosynthesis. Delineating the stages of signal transduction cascades is critical in understanding how processes such as ripening are controlled, with NO presented as a ripening suppressor and inhibitor of ethylene. This study underpins the importance of examining beyond transcriptomic studies and including analysis of protein post-translational modification in uncovering regulatory networks. (Summary by Orla Sherwood, @orlasherwood) New Phytol. 10.1111/nph.18071
Mechanisms of phytohormone modulation and their interface with other regulatory molecules are still being uncovered. Nitric oxide (NO) is a regulatory molecule which has been shown to affect ethylene biosynthesis through the action of ETHYLENE RESPONSE FACTOR 5 (ERF5) in apple fruit. New work by Ji et al. demonstrates that ERF5 nucleocytoplasmic shuttling is dependent on its phosphorylation state. Following application of a NO donor, ERF5 expression increases while ethylene biosynthesis is inhibited. NO stimulates PP2C57-mediated dephosphorylation of ERF5 and facilitates ERF5 translocation from the nucleus to cytoplasm where it suppresses ethylene biosynthesis. Delineating the stages of signal transduction cascades is critical in understanding how processes such as ripening are controlled, with NO presented as a ripening suppressor and inhibitor of ethylene. This study underpins the importance of examining beyond transcriptomic studies and including analysis of protein post-translational modification in uncovering regulatory networks. (Summary by Orla Sherwood, @orlasherwood) New Phytol. 10.1111/nph.18071
Nitrate transporter NRT2.1 is a determinant of a nitrogen acquisition switch between nitrate uptake and symbiosis in Lotus japonicus
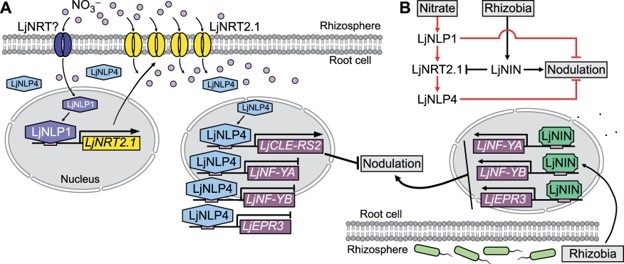 When nitrogen or nitrate availability is high, symbiotic nodule generation is curtailed to conserve the associated energy cost. But how does the presence of nitrate interfere with nodulation and symbiosis? Previously, genetic studies identified nitrate unresponsive symbiosis (nrsym) mutants in Lotus japonicus in which nodule formation is insensitive to nitrate levels. Here, Misawa et al. analyzed two new nrsym mutants, which they found to be affected in the LjNRT2.1 gene that encodes a high affinity nitrate transporter. Further analysis demonstrated the complex interaction between this nitrate transporter and transcription factors previously shown to promote nodulation, such as NODULE INCEPTION (NIN), or suppress nodulation, such as NIN-LIKE PROTEINs (LjNLPs). Briefly, LjNPL4 induces expression of LjNRT2.1, promoting nitrate uptake. The elevated cellular nitrate causes LjNLP4 to move into the nucleus, promoting expression of genes that suppress nodulation. Conversely, in the presence of rhizobia, LjNIN competitively binds to the LjNRT2.1 promoter and suppresses nitrate uptake. Thus, LjNRT2.1 acts as a switch between nitrate uptake and symbiosis. (Summary by Lekshmy Sathee @lekshmysnair) Plant Cell 10.1093/plcell/koac046
When nitrogen or nitrate availability is high, symbiotic nodule generation is curtailed to conserve the associated energy cost. But how does the presence of nitrate interfere with nodulation and symbiosis? Previously, genetic studies identified nitrate unresponsive symbiosis (nrsym) mutants in Lotus japonicus in which nodule formation is insensitive to nitrate levels. Here, Misawa et al. analyzed two new nrsym mutants, which they found to be affected in the LjNRT2.1 gene that encodes a high affinity nitrate transporter. Further analysis demonstrated the complex interaction between this nitrate transporter and transcription factors previously shown to promote nodulation, such as NODULE INCEPTION (NIN), or suppress nodulation, such as NIN-LIKE PROTEINs (LjNLPs). Briefly, LjNPL4 induces expression of LjNRT2.1, promoting nitrate uptake. The elevated cellular nitrate causes LjNLP4 to move into the nucleus, promoting expression of genes that suppress nodulation. Conversely, in the presence of rhizobia, LjNIN competitively binds to the LjNRT2.1 promoter and suppresses nitrate uptake. Thus, LjNRT2.1 acts as a switch between nitrate uptake and symbiosis. (Summary by Lekshmy Sathee @lekshmysnair) Plant Cell 10.1093/plcell/koac046
Arabidopsis CHROMATIN REMODELING 19 acts as a transcriptional repressor and contributes to plant pathogen resistance
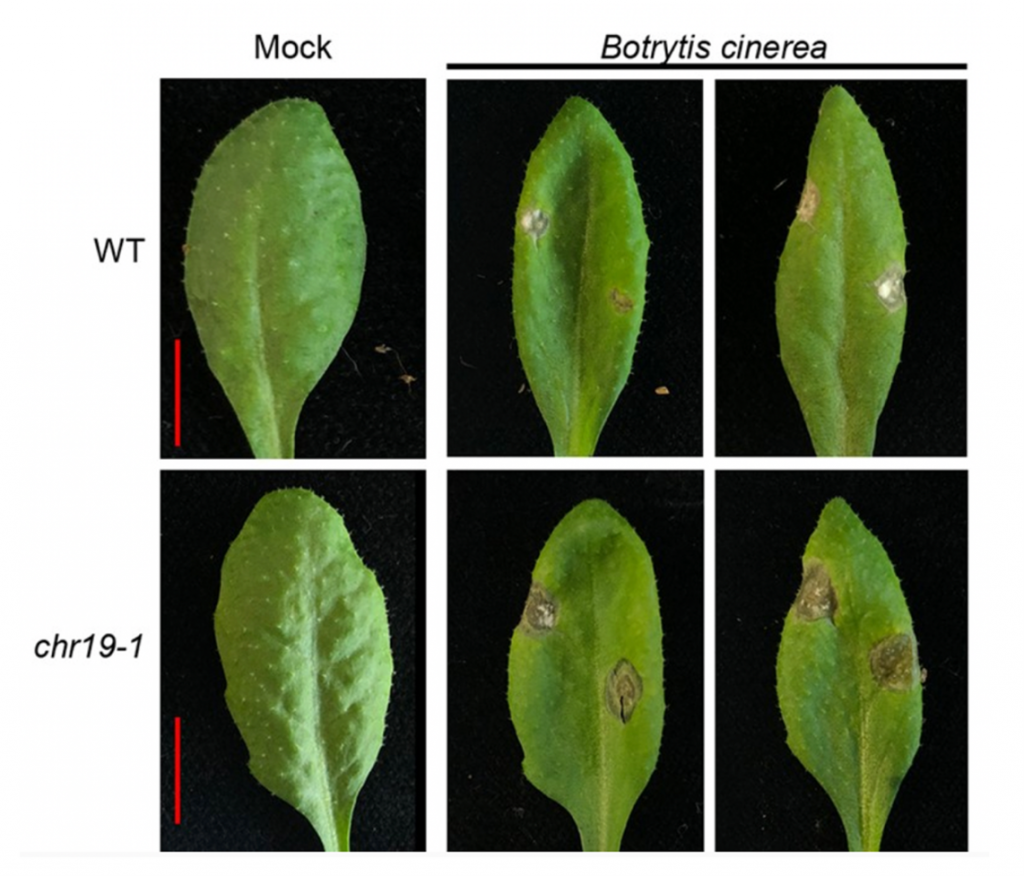 Chromatin remodellers are highly conserved eukaryotic proteins with regulatory roles in various aspects of DNA metabolism including DNA repair, gene expression and mitosis. Here, Kang et al. examine the function of the CHROMATIN REMODELLING19 (CHR19) in Arabidopsis thaliana. Using two T-DNA insertion lines, the researchers first showed that unlike its animal and yeast homologues, CHR19 does not appear to function in DNA repair, with chr19 displaying equal tolerance to DNA damaging agents as wild type plants. RNA-sequencing revealed that a large number of genes are upregulated in the chr19-1 mutant, with genes involved in the salicylic acid and jasmonic acid defence pathways being particularly enriched. To further probe this role of CHR19, the group challenged chr19 plants with bacterial and fungal pathogens. Interestingly the chr19 lines were equally susceptible to the hemibiotrophic bacteria Pseudomonas syringae, but significantly more susceptible to the fungal necrotroph Botrytis cinerea. Finally, the researchers showed that treatment with exogenous salicylic acid reduces the transcript levels of CHR19, and reduces the enrichment of CHR19 at the genomic regions encoding the salicylic acid responsive genes PR1 and WRKY70. Jasmonic acid did not have this effect on CHR19 expression, suggesting that rather than acting as a global suppressor of defence gene expression, CHR19 acts to fine tune specific aspects of plant immunity. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koab318
Chromatin remodellers are highly conserved eukaryotic proteins with regulatory roles in various aspects of DNA metabolism including DNA repair, gene expression and mitosis. Here, Kang et al. examine the function of the CHROMATIN REMODELLING19 (CHR19) in Arabidopsis thaliana. Using two T-DNA insertion lines, the researchers first showed that unlike its animal and yeast homologues, CHR19 does not appear to function in DNA repair, with chr19 displaying equal tolerance to DNA damaging agents as wild type plants. RNA-sequencing revealed that a large number of genes are upregulated in the chr19-1 mutant, with genes involved in the salicylic acid and jasmonic acid defence pathways being particularly enriched. To further probe this role of CHR19, the group challenged chr19 plants with bacterial and fungal pathogens. Interestingly the chr19 lines were equally susceptible to the hemibiotrophic bacteria Pseudomonas syringae, but significantly more susceptible to the fungal necrotroph Botrytis cinerea. Finally, the researchers showed that treatment with exogenous salicylic acid reduces the transcript levels of CHR19, and reduces the enrichment of CHR19 at the genomic regions encoding the salicylic acid responsive genes PR1 and WRKY70. Jasmonic acid did not have this effect on CHR19 expression, suggesting that rather than acting as a global suppressor of defence gene expression, CHR19 acts to fine tune specific aspects of plant immunity. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koab318
Serendipity: Story of breeding dual resistance
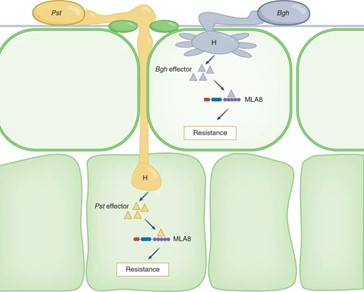 How would you feel if you developed a plant that is tolerant to a particular stress stimulus and found out that it can also tolerate another stress? Bettgenhaeuser and found that this happened when plant breeders developed barley varieties resistant to the powdery mildew disease (caused by the fungal pathogen B. graminis f. sp. hordei; Bgh) and at the same time introduced resistance to wheat stripe rust, caused by a different fungal pathogen. The genetic locus Mildew locus a (Mla) has considerable allelic diversity, and some alleles confer resistance to Bgh. Bettgenhaeuser and colleagues showed that the resistance in domesticated barley towards wheat stripe rust (caused by Puccinia striiformis f. sp. tritici; Pst) is under the control of three quantitative trait loci, designated Rps6, Rps7 and Rps8, and that Rps7 is the major component of resistance in their investigation. Their analysis further showed that the Rps7 and Mla loci are linked, and specifically the Mla8 gene is responsible for conferring resistance to both powdery mildew and stem rust. In an accompanying News and Views, Saur and colleagues provide an excellent illustration to summarise this dual resistance. The MLA8 immune receptor recognizes the Bgh effector AVRA8 to initiate plant defence response, whereas the effectors produced by Pst are currently unknown. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Comms. 10.1038/s41467-021-27288-3 and 10.1038/s41477-022-01097-y
How would you feel if you developed a plant that is tolerant to a particular stress stimulus and found out that it can also tolerate another stress? Bettgenhaeuser and found that this happened when plant breeders developed barley varieties resistant to the powdery mildew disease (caused by the fungal pathogen B. graminis f. sp. hordei; Bgh) and at the same time introduced resistance to wheat stripe rust, caused by a different fungal pathogen. The genetic locus Mildew locus a (Mla) has considerable allelic diversity, and some alleles confer resistance to Bgh. Bettgenhaeuser and colleagues showed that the resistance in domesticated barley towards wheat stripe rust (caused by Puccinia striiformis f. sp. tritici; Pst) is under the control of three quantitative trait loci, designated Rps6, Rps7 and Rps8, and that Rps7 is the major component of resistance in their investigation. Their analysis further showed that the Rps7 and Mla loci are linked, and specifically the Mla8 gene is responsible for conferring resistance to both powdery mildew and stem rust. In an accompanying News and Views, Saur and colleagues provide an excellent illustration to summarise this dual resistance. The MLA8 immune receptor recognizes the Bgh effector AVRA8 to initiate plant defence response, whereas the effectors produced by Pst are currently unknown. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Comms. 10.1038/s41467-021-27288-3 and 10.1038/s41477-022-01097-y
MAP kinase cascade acts as a hub to decide the ways to fight infection
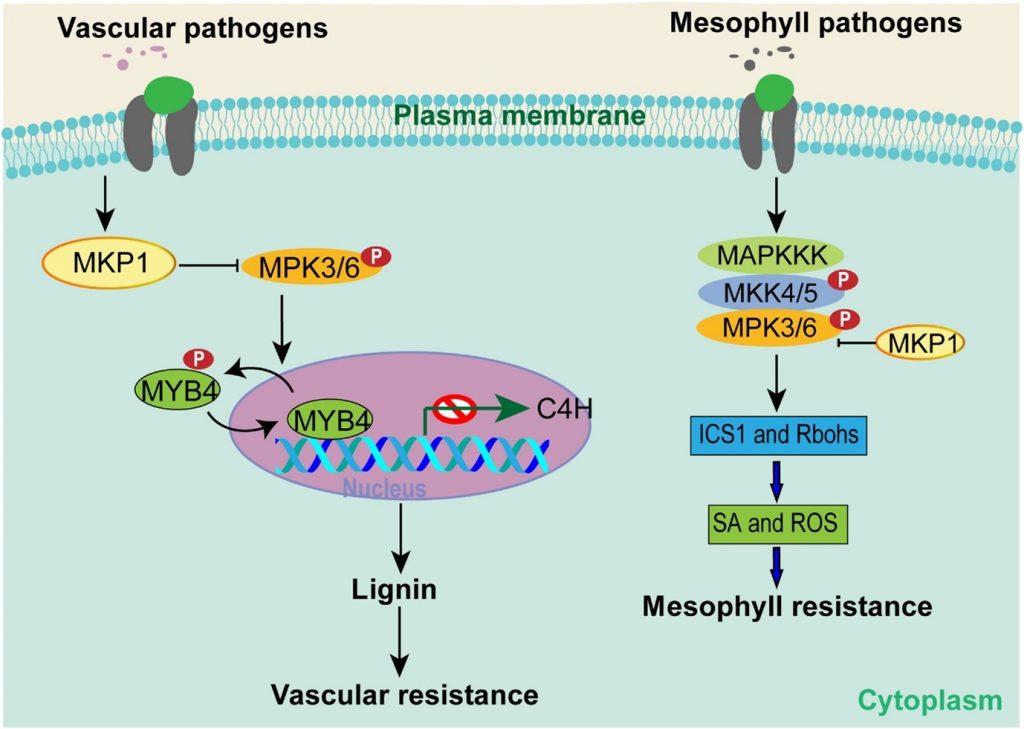 Much of our understanding of plant immunity comes from studies of pathogens that infect mesophyll tissues, (e.g., Pseudomonas syringae). However, there are many pathogens that specifically invade vascular tissues (e.g., Xanthomonas oryzae pv. oryzae; Xoo), which causes rice bacterial blight. In a recent report by Lin et al., the authors detected a signaling cascade that provides specificity between vascular or mesophyll pathogens. This cascade includes a phosphatase MKP1, two well-studied MAP kinases MPK3 and MPK6, and a transcription factor MYB4. The MKP1-MAPK cascade is known to negatively regulate immunity in mesophyll cells by ROS and SA pathways. On the other hand, the authors observed that in vascular tissue MKP1-MAPK signaling induces plant defense by lignin biosynthesis via inactivation of MYB4. This immunity leads to resistance against vascular pathogens in rice and Arabidopsis. Thus, this study reveals how plants have evolved to use the same signaling players in different ways to fight against different types of pathogens. (Summary by Kamal Kumar Malukani, @KamalMalukani). Science Advances 10.1126/sciadv.abg8723.
Much of our understanding of plant immunity comes from studies of pathogens that infect mesophyll tissues, (e.g., Pseudomonas syringae). However, there are many pathogens that specifically invade vascular tissues (e.g., Xanthomonas oryzae pv. oryzae; Xoo), which causes rice bacterial blight. In a recent report by Lin et al., the authors detected a signaling cascade that provides specificity between vascular or mesophyll pathogens. This cascade includes a phosphatase MKP1, two well-studied MAP kinases MPK3 and MPK6, and a transcription factor MYB4. The MKP1-MAPK cascade is known to negatively regulate immunity in mesophyll cells by ROS and SA pathways. On the other hand, the authors observed that in vascular tissue MKP1-MAPK signaling induces plant defense by lignin biosynthesis via inactivation of MYB4. This immunity leads to resistance against vascular pathogens in rice and Arabidopsis. Thus, this study reveals how plants have evolved to use the same signaling players in different ways to fight against different types of pathogens. (Summary by Kamal Kumar Malukani, @KamalMalukani). Science Advances 10.1126/sciadv.abg8723.
Ignoring plant science: When you love the plants but not the plant science
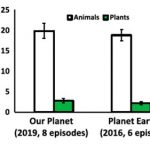
 Have you ever heard the terms ‘zoochauvinism’, ‘plant blindness’ or ‘plant awareness disparity’? Do you know that in a Twitter poll about 30% of the participants voted plants out of wildlife? Ever noticed how important documentaries like ‘Planet Earth II’ and ‘Our Planet’ prefer animals in their content over plants? All these discriminatory and potentially deadly (for plant science) topics are discussed in the Opinion article by Burke, Sherwood and colleagues in Plants People Planet. In the article they demonstrate how society has constantly undervalued plants, and plant science/scientists. However they also discuss trends which show that the appreciation for plants has gone up in recent years; for example, the rise of ‘plantfluencers’ and ‘plantstagrammers,’ or LEGO releasing a Botanical Collection, or the younger generation shifting towards plants over animals as a food source. Perhaps there is still hope for the broader recognition of plants as vital to life. We can contribute our bit as plant scientists by discussing a plant science topic with the public; the authors give an example of mixing a discussion on house plant health with stress physiology. Or, maybe we can start a trend in Twitter to change the name of the channel from ‘Animal Planet’ to ‘Plant and Animal Planet’? (Summary by Sibaji K Sanyal @SibajiSanyal) Plants People Planet 10.1002/ppp3.10257
Have you ever heard the terms ‘zoochauvinism’, ‘plant blindness’ or ‘plant awareness disparity’? Do you know that in a Twitter poll about 30% of the participants voted plants out of wildlife? Ever noticed how important documentaries like ‘Planet Earth II’ and ‘Our Planet’ prefer animals in their content over plants? All these discriminatory and potentially deadly (for plant science) topics are discussed in the Opinion article by Burke, Sherwood and colleagues in Plants People Planet. In the article they demonstrate how society has constantly undervalued plants, and plant science/scientists. However they also discuss trends which show that the appreciation for plants has gone up in recent years; for example, the rise of ‘plantfluencers’ and ‘plantstagrammers,’ or LEGO releasing a Botanical Collection, or the younger generation shifting towards plants over animals as a food source. Perhaps there is still hope for the broader recognition of plants as vital to life. We can contribute our bit as plant scientists by discussing a plant science topic with the public; the authors give an example of mixing a discussion on house plant health with stress physiology. Or, maybe we can start a trend in Twitter to change the name of the channel from ‘Animal Planet’ to ‘Plant and Animal Planet’? (Summary by Sibaji K Sanyal @SibajiSanyal) Plants People Planet 10.1002/ppp3.10257