Plant Science Research Weekly: March 15th
Review: Linking autophagy to abiotic and biotic stress responses ($)
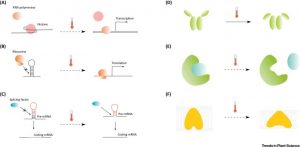
 Autophagy means “self-eating” in ancient Greek. It’s a process in which cellular components are delivered to lytic vacuoles to be reused. This recycling process promotes abiotic and biotic stress tolerance. In this review, Signorelli et al. highlight in detail plant autophagy in abiotic and biotic stress responses and how these responses are triggered to affect autophagy and its signaling pathway. The energy sensors, SNF-related kinase 1 (SnRK1) and target of rapamycin (TOR), as well as abscisic acid (ABA) and its signaling kinase SnRK2 play key roles in the regulation of autophagy under stress conditions. Furthermore, the contribution of reactive oxygen species (ROS) to the establishment of autophagy, the importance of osmotic adjustment, and the role of GABA in plant survival are discussed. Determining the interplay between autophagy and coordinated pathways under various stresses will help researchers to develop novel strategies to improve plant breeding under the threaten of stresses. (Summary by Jin Liao) Trends Plant Sci. 110.1016/j.tplants.2019.02.001
Autophagy means “self-eating” in ancient Greek. It’s a process in which cellular components are delivered to lytic vacuoles to be reused. This recycling process promotes abiotic and biotic stress tolerance. In this review, Signorelli et al. highlight in detail plant autophagy in abiotic and biotic stress responses and how these responses are triggered to affect autophagy and its signaling pathway. The energy sensors, SNF-related kinase 1 (SnRK1) and target of rapamycin (TOR), as well as abscisic acid (ABA) and its signaling kinase SnRK2 play key roles in the regulation of autophagy under stress conditions. Furthermore, the contribution of reactive oxygen species (ROS) to the establishment of autophagy, the importance of osmotic adjustment, and the role of GABA in plant survival are discussed. Determining the interplay between autophagy and coordinated pathways under various stresses will help researchers to develop novel strategies to improve plant breeding under the threaten of stresses. (Summary by Jin Liao) Trends Plant Sci. 110.1016/j.tplants.2019.02.001
Opinion. Feeling the heat: Searching for plant thermosensors
 How does a plant sense the elevation in atmospheric temperature? What are the missing links betweena rise in atmospheric temperature, its sensing, and responses triggered? Vu et al. discuss how conformational and structural changes of DNA, RNA and proteins could be the sensors to temperature change. For example the temperature rise above 22OC indirectly induces the displacement of histone H2A.Z from nucleosomes in promoters and triggers the transcription of genes. The article also illustrates the various modes of temperature sensing by well-known plant development regulators such as FLOWERING LOCUS M (FLM; alternative splicing) and LONG ELONGARE0 HYPOCOTYL (LHY; single nucleotide polymorphism). Conclusively, the authors argued the need for more studies to explore biological thermosensors in plants in light of the expected rise in global temperature and its influence on crop productivity. (Summary by Kaushal Kumar Bhati) Trends Plant Sci 10.1016/j.tplants.2018.11.004
How does a plant sense the elevation in atmospheric temperature? What are the missing links betweena rise in atmospheric temperature, its sensing, and responses triggered? Vu et al. discuss how conformational and structural changes of DNA, RNA and proteins could be the sensors to temperature change. For example the temperature rise above 22OC indirectly induces the displacement of histone H2A.Z from nucleosomes in promoters and triggers the transcription of genes. The article also illustrates the various modes of temperature sensing by well-known plant development regulators such as FLOWERING LOCUS M (FLM; alternative splicing) and LONG ELONGARE0 HYPOCOTYL (LHY; single nucleotide polymorphism). Conclusively, the authors argued the need for more studies to explore biological thermosensors in plants in light of the expected rise in global temperature and its influence on crop productivity. (Summary by Kaushal Kumar Bhati) Trends Plant Sci 10.1016/j.tplants.2018.11.004
A rich evolutionary history of transposable element families in the maize genome
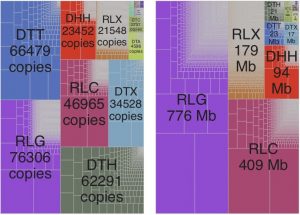 Transposable elements (TEs) make up nearly ~85% of the maize genome. Stitzer et al. report a comprehensive analysis of diverse TE families in maize based on the improved annotation of the updated B73 genome assembly. The authors found all of the known plant TE superfamilies in the maize genome, with Long Terminal Repeats (LTRs) occupying most of the genomic space, and Terminal Inverted Repeats (TIRs) accounting for the highest number of copies. In general, older TE family members coded for proteins whereas younger TE members were noncoding. As expected, most TE families exhibit high levels of DNA methylation, which limit their expression and activity, but a subset of TE families show variable levels and patterns of methylation. The authors attribute such variation as adaptation to the maize genomic environment that each new TE experiences. These TE family annotations, and the associated analysis scripts can be further explored at https://mcstitzer.shinyapps.io/maize_te_families/, and https://github.com/mcstitzer/maize_genomic_ecosystem respectively. (Summary by Saima Shahid) bioRxiv 10.1101/559922.
Transposable elements (TEs) make up nearly ~85% of the maize genome. Stitzer et al. report a comprehensive analysis of diverse TE families in maize based on the improved annotation of the updated B73 genome assembly. The authors found all of the known plant TE superfamilies in the maize genome, with Long Terminal Repeats (LTRs) occupying most of the genomic space, and Terminal Inverted Repeats (TIRs) accounting for the highest number of copies. In general, older TE family members coded for proteins whereas younger TE members were noncoding. As expected, most TE families exhibit high levels of DNA methylation, which limit their expression and activity, but a subset of TE families show variable levels and patterns of methylation. The authors attribute such variation as adaptation to the maize genomic environment that each new TE experiences. These TE family annotations, and the associated analysis scripts can be further explored at https://mcstitzer.shinyapps.io/maize_te_families/, and https://github.com/mcstitzer/maize_genomic_ecosystem respectively. (Summary by Saima Shahid) bioRxiv 10.1101/559922.
Ubiquitin-dependent chloroplast-associated protein degradation in plants
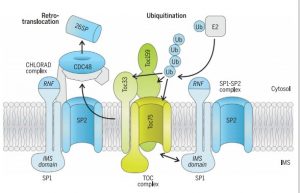 The fidelity of the chloroplast proteome is a major factor in the functional efficiency of photosynthesis. A RING-type ubiquitin E3 ligase, SP1, is an outer envelope-localized protein in the chloroplast which ubiquitinates the protein import translocases (TOC proteins) and thus target them to 26S mediated degradation. In recent work, Ling et al. identified a mutation that suppresses the chlorosis phenotype of the Toc33 mutant (ppil), which they named suppressor of ppi1 locus 2 (sp2). Using genetic and protein-interaction methods, the authors confirmed that AAA+ chaperone CDC48 and SP2 protein participate in the same proteolytic pathway with SP1. Thus, they conceptualize the multicomponent system for chloroplast envelope protein removal by chloroplast-associated protein degradation (CHLORAD). (Summary by Kaushal Kumar Bhati) Science 10.1126/science.aav4467
The fidelity of the chloroplast proteome is a major factor in the functional efficiency of photosynthesis. A RING-type ubiquitin E3 ligase, SP1, is an outer envelope-localized protein in the chloroplast which ubiquitinates the protein import translocases (TOC proteins) and thus target them to 26S mediated degradation. In recent work, Ling et al. identified a mutation that suppresses the chlorosis phenotype of the Toc33 mutant (ppil), which they named suppressor of ppi1 locus 2 (sp2). Using genetic and protein-interaction methods, the authors confirmed that AAA+ chaperone CDC48 and SP2 protein participate in the same proteolytic pathway with SP1. Thus, they conceptualize the multicomponent system for chloroplast envelope protein removal by chloroplast-associated protein degradation (CHLORAD). (Summary by Kaushal Kumar Bhati) Science 10.1126/science.aav4467
Complete biosynthesis of cannabinoids and their unnatural analogues in yeast ($)
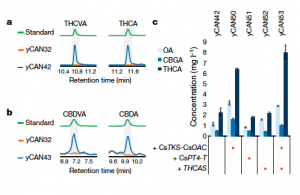 Cannabis sativa L. produces cannabinoid compounds that interest the medical community for the treatment of symptoms that may be difficult to suppress with common medical treatments. Research on cannabinoids is limited by their low abundance in plant tissue and structurally complex chemical synthesis. Luo et al. addresses these issues through the assembly of a cannabinoid synthesis pathway in Saccharomyces cerevisiae fed with galactose. In this heterologous system, they assembled enzyme pathways involved in the synthesis of key intermediate compounds such as hexanoyl-CoA, olivetolic acid, and cannabigerolic acid, which is a precursor to D9-tetrahydrocannabinolic acid and cannabidiolic acid. Additionally, the authors identify key cannabinoid analogs that can be synthesized by selective feeding of these lines of S. cerevisiae with various fatty acids. This research may help to boost cannabinoid research through increased synthesis of natural compounds for functional studies, as well as aid in the identification of new compounds through investigation of the fatty-acid derived analogs. Summary by Nathan Scinto-Madonich Nature. 10.1038/s41586-019-0978-9
Cannabis sativa L. produces cannabinoid compounds that interest the medical community for the treatment of symptoms that may be difficult to suppress with common medical treatments. Research on cannabinoids is limited by their low abundance in plant tissue and structurally complex chemical synthesis. Luo et al. addresses these issues through the assembly of a cannabinoid synthesis pathway in Saccharomyces cerevisiae fed with galactose. In this heterologous system, they assembled enzyme pathways involved in the synthesis of key intermediate compounds such as hexanoyl-CoA, olivetolic acid, and cannabigerolic acid, which is a precursor to D9-tetrahydrocannabinolic acid and cannabidiolic acid. Additionally, the authors identify key cannabinoid analogs that can be synthesized by selective feeding of these lines of S. cerevisiae with various fatty acids. This research may help to boost cannabinoid research through increased synthesis of natural compounds for functional studies, as well as aid in the identification of new compounds through investigation of the fatty-acid derived analogs. Summary by Nathan Scinto-Madonich Nature. 10.1038/s41586-019-0978-9
The phosphorylation and protein interaction landscape of the plant TOR kinase ($)
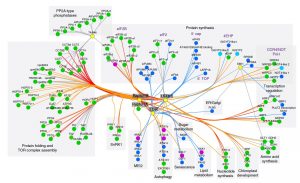 The TOR (target of rapamycin) is a conserved regulator of cellular homeostasis and energy status in several clades of life. TOR signaling is well studied in animals while precise regulation and downstream targets of TOR kinase are not very well explored in plants. Some well-studied TOR kinase targets in plants includes S6 kinase, TAP46 and eIF-4E binding protein. In plants the activity of TOR is regulated by hormones and the SnRK1 complex. Mutant analysis and -omics studies have linked plant TOR signaling to nutrient status, meristem development in seedling and autophagy. Using an extensive phosphor-proteome analysis, Leene et al. have been able to develop a new protein interactome map for up/downstream components of TOR signaling in Arabidopsis. The differential phosphor-proteome analysis resulted in the identification of 83 TOR regulated proteins. Some interesting biological pathways for TOR targets are sugar protein homeostasis, protein folding, lipid and nucleotide metabolism and autophagy. (Summary by Kaushal Kumar Bhati) Nature Plants 10.1038/s41477-019-0378-z
The TOR (target of rapamycin) is a conserved regulator of cellular homeostasis and energy status in several clades of life. TOR signaling is well studied in animals while precise regulation and downstream targets of TOR kinase are not very well explored in plants. Some well-studied TOR kinase targets in plants includes S6 kinase, TAP46 and eIF-4E binding protein. In plants the activity of TOR is regulated by hormones and the SnRK1 complex. Mutant analysis and -omics studies have linked plant TOR signaling to nutrient status, meristem development in seedling and autophagy. Using an extensive phosphor-proteome analysis, Leene et al. have been able to develop a new protein interactome map for up/downstream components of TOR signaling in Arabidopsis. The differential phosphor-proteome analysis resulted in the identification of 83 TOR regulated proteins. Some interesting biological pathways for TOR targets are sugar protein homeostasis, protein folding, lipid and nucleotide metabolism and autophagy. (Summary by Kaushal Kumar Bhati) Nature Plants 10.1038/s41477-019-0378-z
Multiple transcriptional factors control stomata development in rice ($)
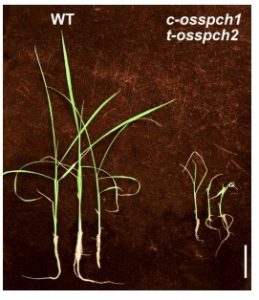 In the model plant Arabidopsis, stomatal development occurs in a scattered pattern with that avoids direct contact between them. In rice and other monocots, the stomata are distributed in a uniformly oriented manner. Such unique patterning and morphology of stomata intrigued Wu et al. to examine if the transcriptional factors (TFs) identified in Arabidopsis share similar function in rice. In Arabidopsis the main bHLH transcriptional factors SPCH (SPEECHLESS), MUTE and FAMA promote stem cell identity to asymmetric cell division leading to stomata and pavement cell formation. Using T-DNA insertion alleles and gene editing, similar functions for these TFs have been observed in rice as in Arabidopsis. In addition the authors identified new roles for SHR (SHORTROOT) and SCR (SCARECROW) in stomata development. (Summary by Suresh Damodaran) New Phytol. 10.1111/nph.15766
In the model plant Arabidopsis, stomatal development occurs in a scattered pattern with that avoids direct contact between them. In rice and other monocots, the stomata are distributed in a uniformly oriented manner. Such unique patterning and morphology of stomata intrigued Wu et al. to examine if the transcriptional factors (TFs) identified in Arabidopsis share similar function in rice. In Arabidopsis the main bHLH transcriptional factors SPCH (SPEECHLESS), MUTE and FAMA promote stem cell identity to asymmetric cell division leading to stomata and pavement cell formation. Using T-DNA insertion alleles and gene editing, similar functions for these TFs have been observed in rice as in Arabidopsis. In addition the authors identified new roles for SHR (SHORTROOT) and SCR (SCARECROW) in stomata development. (Summary by Suresh Damodaran) New Phytol. 10.1111/nph.15766
VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures
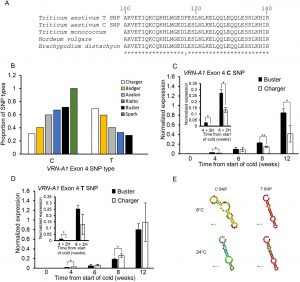 In bread wheat, the VERNALIZATION 1 (VRN1) gene is induced after cold exposure and short days, promoting flowering. In some species the vernalization effect of cold temperature can be lost or reversed after a short exposure to a high temperature (∼35°C); this process is referred as de-vernalization. Very little is known about this response or the genetic regulation under variable growth temperatures. Dixon and colleagues exposed 98 bread wheat winter and spring varieties to two different growing temperature conditions (low, LT, and high, HT, temperature) after vernalization. The response was varied; all but one flowered faster under HT. Charger, a winter variety, flowered earlier under LT. Further analysis showed that Charger has a high copy number of VRN-A1 (VRN1 copy in genome A) and the T-type allele was more frequent in Charger compared to the other winter species analyzed. They also found that genes that repress flowering like VRN2 are re-activated under HT during and after vernalization, and this re-activation depended on photoperiod. The results show how the absence of warm is critical for the completion of vernalization but also that the known temperatures for vernalization could be higher than previously thought. (Summary by Cecilia Vasquez-Robinet) Development 10.1242/dev.172684
In bread wheat, the VERNALIZATION 1 (VRN1) gene is induced after cold exposure and short days, promoting flowering. In some species the vernalization effect of cold temperature can be lost or reversed after a short exposure to a high temperature (∼35°C); this process is referred as de-vernalization. Very little is known about this response or the genetic regulation under variable growth temperatures. Dixon and colleagues exposed 98 bread wheat winter and spring varieties to two different growing temperature conditions (low, LT, and high, HT, temperature) after vernalization. The response was varied; all but one flowered faster under HT. Charger, a winter variety, flowered earlier under LT. Further analysis showed that Charger has a high copy number of VRN-A1 (VRN1 copy in genome A) and the T-type allele was more frequent in Charger compared to the other winter species analyzed. They also found that genes that repress flowering like VRN2 are re-activated under HT during and after vernalization, and this re-activation depended on photoperiod. The results show how the absence of warm is critical for the completion of vernalization but also that the known temperatures for vernalization could be higher than previously thought. (Summary by Cecilia Vasquez-Robinet) Development 10.1242/dev.172684
De novo origins of multicellularity in response to predation
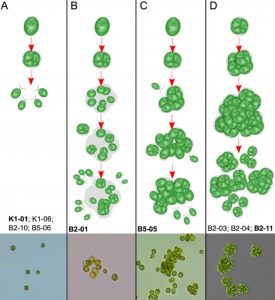 Multicellularity has evolved independently many times in eukaryotes (e.g. land plants, fungi, animals). Predation is one driver that is hypothesised to lead to this evolution from unicellular ancestors. Here, using evolution experiments, Herron et al. demonstrate that the unicellular green algae Chlamydomonas reinhardtii develops multicellular structures under selective pressure of the predatory filter feeder Paramecium tetraurelia. These developments that evolved over approximately 750 generations are shown to be heritable after four years in the absence of a predator. The multicellular phenotype provides a fitness benefit because these algae increase their body size becoming less susceptible to predation. Comparisons with similar studies in other unicellular organisms indicates that predation is an ecologically viable selective pressure that could explain the origins of multicellularity (Summary by Alex Bowles) Scientific reports 10.1038/s41598-019-39558-8.
Multicellularity has evolved independently many times in eukaryotes (e.g. land plants, fungi, animals). Predation is one driver that is hypothesised to lead to this evolution from unicellular ancestors. Here, using evolution experiments, Herron et al. demonstrate that the unicellular green algae Chlamydomonas reinhardtii develops multicellular structures under selective pressure of the predatory filter feeder Paramecium tetraurelia. These developments that evolved over approximately 750 generations are shown to be heritable after four years in the absence of a predator. The multicellular phenotype provides a fitness benefit because these algae increase their body size becoming less susceptible to predation. Comparisons with similar studies in other unicellular organisms indicates that predation is an ecologically viable selective pressure that could explain the origins of multicellularity (Summary by Alex Bowles) Scientific reports 10.1038/s41598-019-39558-8.
Independent regulation of symbiotic nodulation by the SUNN negative and CRA2 positive systemic pathways ($)
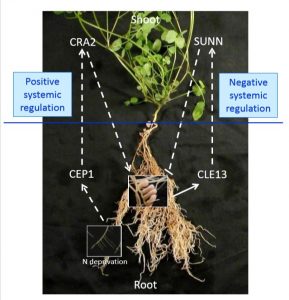 Autoregulation of nodule development (AON) is a process by which leguminous plants control nodule development. SUNN, a LRR-RLK, and a systemic CLE peptide play a negative role in nodulation through systemic signaling. In this paper Laffont et al. showed the another systemic regulation of nodulation in Medicago by CRA2 (COMPACT ROOR ARCHITECTURE 2), a leucine-rich receptor-like kinase (LRR-RLK). Moreover, the nodulation inhibitory effect of MtCLE13 peptide is unaltered in cra2 mutants suggesting an independent pathway of the SUNN-CLE mechanism. It has been demonstrated using grafting studies with different combinations of Wt, sunn, cra2 mutants that a balance in nodulation, maintained by positive systemic regulation by CRA2 and negative systemic regulation by SUNN, is necessary to achieve proper nodulation. (Summary by Suresh Damodaran) Plant Physiol. 10.1104/pp.18.01588
Autoregulation of nodule development (AON) is a process by which leguminous plants control nodule development. SUNN, a LRR-RLK, and a systemic CLE peptide play a negative role in nodulation through systemic signaling. In this paper Laffont et al. showed the another systemic regulation of nodulation in Medicago by CRA2 (COMPACT ROOR ARCHITECTURE 2), a leucine-rich receptor-like kinase (LRR-RLK). Moreover, the nodulation inhibitory effect of MtCLE13 peptide is unaltered in cra2 mutants suggesting an independent pathway of the SUNN-CLE mechanism. It has been demonstrated using grafting studies with different combinations of Wt, sunn, cra2 mutants that a balance in nodulation, maintained by positive systemic regulation by CRA2 and negative systemic regulation by SUNN, is necessary to achieve proper nodulation. (Summary by Suresh Damodaran) Plant Physiol. 10.1104/pp.18.01588
BSL family members are employed by pathogen as “moles” in S. tuberosum
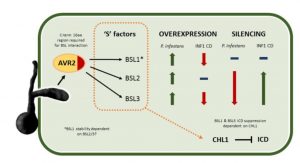 To secure food availability for the world population, plant scientists study the interactions between crop species and pathogens. Plants have robust defense strategies that they employ when attacked. On the other hand, microbes have multiple strategies to manipulate host functions to suppress these defenses and/or enhance the growth of the microbe. Recently Turnbull et al. found that in S. tuberosum, Phytophthora infestans effector PiAVR2 could interact with all three BSU1-like (BSL) family members. In this way pathogens exploit growth/immune crosstalk by upregulating BR signaling to attenuate the immune response. All three BSL proteins act as “moles” (or susceptibility factors) in P. infestans infection, allowing increased disease susceptibility when overexpressed and decreased disease progression when silenced. This study is interesting as it shows that a pathogen can exploit the crosstalk between BR pathway and immune signaling. (Summary by Nanxun Qin). Plant Physiol. 10.1104/pp.18.01143
To secure food availability for the world population, plant scientists study the interactions between crop species and pathogens. Plants have robust defense strategies that they employ when attacked. On the other hand, microbes have multiple strategies to manipulate host functions to suppress these defenses and/or enhance the growth of the microbe. Recently Turnbull et al. found that in S. tuberosum, Phytophthora infestans effector PiAVR2 could interact with all three BSU1-like (BSL) family members. In this way pathogens exploit growth/immune crosstalk by upregulating BR signaling to attenuate the immune response. All three BSL proteins act as “moles” (or susceptibility factors) in P. infestans infection, allowing increased disease susceptibility when overexpressed and decreased disease progression when silenced. This study is interesting as it shows that a pathogen can exploit the crosstalk between BR pathway and immune signaling. (Summary by Nanxun Qin). Plant Physiol. 10.1104/pp.18.01143
Century-old museum specimens predict a timeline for declines in monarch butterflies and their host milkweeds
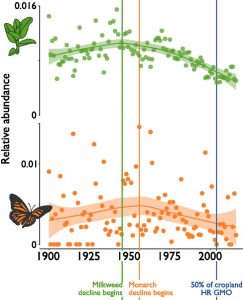 Milkweeds are often found in agriculture fields, and are susceptible to herbicides sprayed in such fields. A decline in milkweeds, which provide food for monarch butterflies, has been historically linked to the wide cultivation of herbicide-resistant genetically modified (GM) crops. Boyle et al. attempt to reconstruct a timeline of monarch and milkweed abundance based on museum and herbaria specimens collected over a century (1900 – 2016). Based on their analyses, the authors conclude that milkweed and monarch populations started to decrease around 1950, decades before the introduction of genetically modified crops. The authors hypothesize that multiple events such as merging of smaller farms into larger farms and increased herbicide use might have triggered the decline of monarch butterfly instead of a single event, i.e., the use of GM crops. The authors clearly articulate potential biases and limitations in estimating insect abundance based on museum and herbaria collections. Nevertheless, this study provides an interesting approach based on systematic analysis of collection data for predicting broad trends in host-insect dynamics over long periods. (Summary by Saima Shahid) PNAS 10.1073/pnas.1811437116.
Milkweeds are often found in agriculture fields, and are susceptible to herbicides sprayed in such fields. A decline in milkweeds, which provide food for monarch butterflies, has been historically linked to the wide cultivation of herbicide-resistant genetically modified (GM) crops. Boyle et al. attempt to reconstruct a timeline of monarch and milkweed abundance based on museum and herbaria specimens collected over a century (1900 – 2016). Based on their analyses, the authors conclude that milkweed and monarch populations started to decrease around 1950, decades before the introduction of genetically modified crops. The authors hypothesize that multiple events such as merging of smaller farms into larger farms and increased herbicide use might have triggered the decline of monarch butterfly instead of a single event, i.e., the use of GM crops. The authors clearly articulate potential biases and limitations in estimating insect abundance based on museum and herbaria collections. Nevertheless, this study provides an interesting approach based on systematic analysis of collection data for predicting broad trends in host-insect dynamics over long periods. (Summary by Saima Shahid) PNAS 10.1073/pnas.1811437116.