Plant Science Research Weekly: March 15, 2024
Review: Synthetic algal genomes
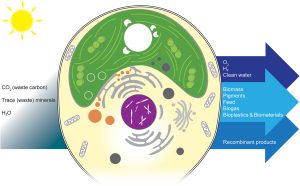 As photosynthetic organisms, algae can harness solar energy to do useful things, from environmental cleanup to producing fuels and other beneficial molecules. This review by Goold et al. provides an overview of how algae can be valuable platforms for synthetic biology and metabolic engineering through the incorporation of synthetic genomes. The authors build on the work of the Synthetic Yeast Genome Project (Sc2.0), which endeavors to redesign the yeast genome to make it more concise and easier to manipulate (for example, by introducing recombination sites and adding “synthetic supernumerary neochromosomes”); basically, if you want to use a cell as a factory, start by tidying up the junk and making it into an environment designed for efficiency. The best system for a synthetic algal genome project isn’t quite as clearcut as yeast. The authors first look at the potential of Chlamydomonas reinhardtii, the algal superstar. However good it is as a model system, it isn’t optimal for this approach due to its large genome, so they then discuss other algal species as candidates for synthetic genomes. Beneficial traits include rapid growth, both diploid and haploid states with the possibility of sexual reproduction, interesting metabolic pathways, and the ability to grow heterotrophically, particularly when photosynthetic genes are being manipulated. Given the great diversity of algae, there are several good candidates and it seems likely that as this technology develops many will be employed. This is a very thought-provoking article that provides a peek into the future of algal biotechnology. (Summary by Mary Williams @PlantTeaching) Cell Genomics 10.1016/j.xgen.2024.100505
As photosynthetic organisms, algae can harness solar energy to do useful things, from environmental cleanup to producing fuels and other beneficial molecules. This review by Goold et al. provides an overview of how algae can be valuable platforms for synthetic biology and metabolic engineering through the incorporation of synthetic genomes. The authors build on the work of the Synthetic Yeast Genome Project (Sc2.0), which endeavors to redesign the yeast genome to make it more concise and easier to manipulate (for example, by introducing recombination sites and adding “synthetic supernumerary neochromosomes”); basically, if you want to use a cell as a factory, start by tidying up the junk and making it into an environment designed for efficiency. The best system for a synthetic algal genome project isn’t quite as clearcut as yeast. The authors first look at the potential of Chlamydomonas reinhardtii, the algal superstar. However good it is as a model system, it isn’t optimal for this approach due to its large genome, so they then discuss other algal species as candidates for synthetic genomes. Beneficial traits include rapid growth, both diploid and haploid states with the possibility of sexual reproduction, interesting metabolic pathways, and the ability to grow heterotrophically, particularly when photosynthetic genes are being manipulated. Given the great diversity of algae, there are several good candidates and it seems likely that as this technology develops many will be employed. This is a very thought-provoking article that provides a peek into the future of algal biotechnology. (Summary by Mary Williams @PlantTeaching) Cell Genomics 10.1016/j.xgen.2024.100505
Review: Tapping onto an integrative framework for successful deployment of microbiomes in agriculture
 This review by Berruto and Demirer calls for an interdisciplinary approach at the interface of experimental and computational frameworks to address the long-standing challenge of field application of beneficial soil microbiomes. This challenge persists due to our limited understanding of the complex principles governing plant-microbe and microbe-microbe interactions. The authors provide a comprehensive overview of both plant and microbial factors that determine this plant-microbe detente and proposed strategies to design microbial consortia for predictable plant phenotypes. Combining genome-wide association studies (GWAS) of host plants with metagenome-wide association studies (MWAS) could reveal target loci for plant breeding and improved plant-microbe associations. The authors propose a shift in our approach to integrating such datasets by considering feature interactions of both host and microbiome, especially through use of machine learning (ML). Conversely, the complexity of microbial interactions (pairwise or higher order) and their emergent properties could be mapped by developing ML based predictive frameworks incorporating data from co-inoculation experiments (putting two strains together and assessing plant phenotypes) with genome scale metabolic models (GSMMs). Finally, these strategies could be supplemented with emerging engineering techniques for both the host plant and its microbiome to precisely manipulate plant phenotypes. (Summary by Arijit Mukherjee @ArijitM61745830) Trends Microbiol. https://doi.org/10.1016/j.tim.2024.02.003
This review by Berruto and Demirer calls for an interdisciplinary approach at the interface of experimental and computational frameworks to address the long-standing challenge of field application of beneficial soil microbiomes. This challenge persists due to our limited understanding of the complex principles governing plant-microbe and microbe-microbe interactions. The authors provide a comprehensive overview of both plant and microbial factors that determine this plant-microbe detente and proposed strategies to design microbial consortia for predictable plant phenotypes. Combining genome-wide association studies (GWAS) of host plants with metagenome-wide association studies (MWAS) could reveal target loci for plant breeding and improved plant-microbe associations. The authors propose a shift in our approach to integrating such datasets by considering feature interactions of both host and microbiome, especially through use of machine learning (ML). Conversely, the complexity of microbial interactions (pairwise or higher order) and their emergent properties could be mapped by developing ML based predictive frameworks incorporating data from co-inoculation experiments (putting two strains together and assessing plant phenotypes) with genome scale metabolic models (GSMMs). Finally, these strategies could be supplemented with emerging engineering techniques for both the host plant and its microbiome to precisely manipulate plant phenotypes. (Summary by Arijit Mukherjee @ArijitM61745830) Trends Microbiol. https://doi.org/10.1016/j.tim.2024.02.003
Review: Deep learning in image-based plant phenotyping
 As a writer and an editor, I an horrified by the idea that thinking can be replaced by artificial intelligence. But I do recognize that deep learning / machine learning / artificial intelligence can provide major opportunities for data analysis, as eloquently described in this review article by Murphy et al. that focuses on the applications of deep learning for plant image data and phenotyping. They start off with some clear definitions (including helpful diagrams) of the terminology employed, whether about computer vision and image segmentation or different types of deep learning and neural networks. They next discuss best practices, emphasizing the importance of carefully selecting the training and validation datasets, and define the different metrics that can be used to assess the success of the deep learning model. They wrap up with some examples of how deep learning can be used in plant classification (including weed versus non-weed) and phenomics, including early detection of stress or disease, and a call for more training in these methods. Like it or not, machine learning is part of our lives now, and this review is an excellent way to start to appreciate how it can make our science more efficient and effective. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-070523-042828
As a writer and an editor, I an horrified by the idea that thinking can be replaced by artificial intelligence. But I do recognize that deep learning / machine learning / artificial intelligence can provide major opportunities for data analysis, as eloquently described in this review article by Murphy et al. that focuses on the applications of deep learning for plant image data and phenotyping. They start off with some clear definitions (including helpful diagrams) of the terminology employed, whether about computer vision and image segmentation or different types of deep learning and neural networks. They next discuss best practices, emphasizing the importance of carefully selecting the training and validation datasets, and define the different metrics that can be used to assess the success of the deep learning model. They wrap up with some examples of how deep learning can be used in plant classification (including weed versus non-weed) and phenomics, including early detection of stress or disease, and a call for more training in these methods. Like it or not, machine learning is part of our lives now, and this review is an excellent way to start to appreciate how it can make our science more efficient and effective. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-070523-042828
Understanding the role of metabolites in the evolution of C4 photosynthesis
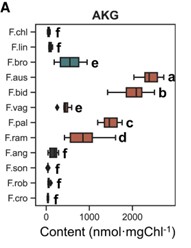 Plants that use C4 photosynthesis, whereby carbon is concentrated around RuBisCo in bundle sheath cells, have high water and nitrogen use efficiencies. Thus, understanding how C4 photosynthesis evolves is of much interest. The Flaveria genus provides a unique opportunity for this, as it contains species that are classed as C3, C4, C4-like and C3-C4 intermediates. Here, Tang et al. grew 12 Flaveria species and used high-performance liquid chromatography–tandem mass spectrometry to measure the concentrations of 40 primary metabolites. Interestingly, α-ketoglutarate concentrations, when normalised to account for different chlorophyll contents, were low in C3 species, but were nearly 1000 times higher in the C3-C4 intermediate F. ramossissima, and higher again in the C4 species F. bidentis. To test whether increased α-ketoglutarate influences photosynthesis, the authors exogenously fed α-ketoglutarate to F. ramossissima petioles and saw an increase in maximum photosynthetic rate. Repeating the same experiment with a pulse of 13C labelled carbon dioxide, revealed there was faster accumulation of oxaloacetate and pyruvate, which are key C4 metabolites. Hence, increasing α-ketoglutarate concentration may be a key step in the transition to C4 photosynthesis. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiae077
Plants that use C4 photosynthesis, whereby carbon is concentrated around RuBisCo in bundle sheath cells, have high water and nitrogen use efficiencies. Thus, understanding how C4 photosynthesis evolves is of much interest. The Flaveria genus provides a unique opportunity for this, as it contains species that are classed as C3, C4, C4-like and C3-C4 intermediates. Here, Tang et al. grew 12 Flaveria species and used high-performance liquid chromatography–tandem mass spectrometry to measure the concentrations of 40 primary metabolites. Interestingly, α-ketoglutarate concentrations, when normalised to account for different chlorophyll contents, were low in C3 species, but were nearly 1000 times higher in the C3-C4 intermediate F. ramossissima, and higher again in the C4 species F. bidentis. To test whether increased α-ketoglutarate influences photosynthesis, the authors exogenously fed α-ketoglutarate to F. ramossissima petioles and saw an increase in maximum photosynthetic rate. Repeating the same experiment with a pulse of 13C labelled carbon dioxide, revealed there was faster accumulation of oxaloacetate and pyruvate, which are key C4 metabolites. Hence, increasing α-ketoglutarate concentration may be a key step in the transition to C4 photosynthesis. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiae077
Functional insights into the TPLATE complex: A key player in clathrin-mediated endocytosis in plant cells
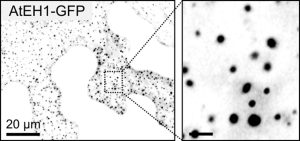 Transporting cargo within a cell may seem straightforward, but it is actually an intricate task. At the plasma membrane in plant cells, this complexity is navigated by a process known as clathrin-mediated endocytosis. Central to this process are two key players: adapter protein AP-2 and the TSET/TPLATE complex (TPC). As an evolutionarily ancient structure, the TPC has emerged as a vital component in this cellular machinery. Acting as an adapter module, it plays a crucial role in shaping the membrane during endocytosis and has been found to interact with ubiquitinated cargo. Structurally, it resembles both COPI and AP complexes; however, the TPC forms an octameric complex and is present during the earliest phases of endocytosis until vesicle scission and uncoating. In Dragwidge et al., the authors delved into the roles of two highly disordered TPC subunits, EH1 and EH2. They discovered that these subunits serve as scaffolds for biomolecular condensation of the complex, a process crucial for influencing endocytosis dynamics as well as recruiting other endocytic proteins. Specifically, the EH domain in EH1 and EH2 was identified as responsible for lipid binding, enabling interaction with the plasma membrane by binding anionic phospholipids. In contrast, the intrinsically disordered region1 (IDR1) affects condensate properties. The authors provide stunning high-resolution microscopic images reinforcing their findings. Their work emphasizes the significant role of condensation and phase separation in the assembly of clathrin-coated vesicles and the recruitment of other accessory proteins at the plasma membrane. (Summary by Ann-Kathrin Rößling @AK_Roessling) 10.1038/s41556-024-01354-6
Transporting cargo within a cell may seem straightforward, but it is actually an intricate task. At the plasma membrane in plant cells, this complexity is navigated by a process known as clathrin-mediated endocytosis. Central to this process are two key players: adapter protein AP-2 and the TSET/TPLATE complex (TPC). As an evolutionarily ancient structure, the TPC has emerged as a vital component in this cellular machinery. Acting as an adapter module, it plays a crucial role in shaping the membrane during endocytosis and has been found to interact with ubiquitinated cargo. Structurally, it resembles both COPI and AP complexes; however, the TPC forms an octameric complex and is present during the earliest phases of endocytosis until vesicle scission and uncoating. In Dragwidge et al., the authors delved into the roles of two highly disordered TPC subunits, EH1 and EH2. They discovered that these subunits serve as scaffolds for biomolecular condensation of the complex, a process crucial for influencing endocytosis dynamics as well as recruiting other endocytic proteins. Specifically, the EH domain in EH1 and EH2 was identified as responsible for lipid binding, enabling interaction with the plasma membrane by binding anionic phospholipids. In contrast, the intrinsically disordered region1 (IDR1) affects condensate properties. The authors provide stunning high-resolution microscopic images reinforcing their findings. Their work emphasizes the significant role of condensation and phase separation in the assembly of clathrin-coated vesicles and the recruitment of other accessory proteins at the plasma membrane. (Summary by Ann-Kathrin Rößling @AK_Roessling) 10.1038/s41556-024-01354-6
Every pair is special!
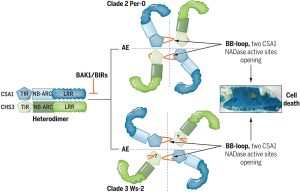 The identification and characterization of resistosome complexes, formed by the oligomerization of intracellular nucleotide-binding leucine-rich repeat receptors (NLRs), has recently been a major focus in the field of plant immunity. Some NLR genes occur as head-to-head pairs encoding proteins that function as heterodimers. Yang et al. investigated the oligomerization requirements for the paired class of NLRs using the Arabidopsis CHS3-CSA1 pair. Using blue native-PAGE and co-IPs, the authors show that clades 2 and 3 of CHS3-CSA1 form heterodimer structures. They further show that upon activation (for example upon effector binding), these heterodimers further oligomerize to form heterotetrameric complexes. Through a series of mutational experiments, the authors further identify specific domain features required for the function and oligomerization of these complexes. Finally, building upon previous studies linking CHS3-CSA1 to the immune co-receptors BAK1 and/or BIRs, the group shows that these co-receptors act as negative regulators of CHS3-CSA1 and inhibit their further oligomerization to the heterotetrameric state. The authors close by highlighting how the requirements they identified for the function and oligomerizing of CHS3-CSA1 differ significantly from another well-studied NLR pair, RRS1-RPS4. These findings serve as an important reminder that overgeneralizations and predictions based off limited representative structures may not capture significant differences in paired NLR structure. (Summary by Tamar Av-Shalom @TamarAvShalom) Science. 10.1126/science.adk3468
The identification and characterization of resistosome complexes, formed by the oligomerization of intracellular nucleotide-binding leucine-rich repeat receptors (NLRs), has recently been a major focus in the field of plant immunity. Some NLR genes occur as head-to-head pairs encoding proteins that function as heterodimers. Yang et al. investigated the oligomerization requirements for the paired class of NLRs using the Arabidopsis CHS3-CSA1 pair. Using blue native-PAGE and co-IPs, the authors show that clades 2 and 3 of CHS3-CSA1 form heterodimer structures. They further show that upon activation (for example upon effector binding), these heterodimers further oligomerize to form heterotetrameric complexes. Through a series of mutational experiments, the authors further identify specific domain features required for the function and oligomerization of these complexes. Finally, building upon previous studies linking CHS3-CSA1 to the immune co-receptors BAK1 and/or BIRs, the group shows that these co-receptors act as negative regulators of CHS3-CSA1 and inhibit their further oligomerization to the heterotetrameric state. The authors close by highlighting how the requirements they identified for the function and oligomerizing of CHS3-CSA1 differ significantly from another well-studied NLR pair, RRS1-RPS4. These findings serve as an important reminder that overgeneralizations and predictions based off limited representative structures may not capture significant differences in paired NLR structure. (Summary by Tamar Av-Shalom @TamarAvShalom) Science. 10.1126/science.adk3468
A diffusible small-RNA-based Turing system dynamically coordinates organ polarity
 The establishment of adaxial-abaxial polarity during the growth of the leaf primordium is a prerequisite for the formation of the flat, thin leaves observed in most (but not all) plants. The micro-RNAs tasiARF and miR165/6, which form concentration gradients within leaf primordia, are major contributors to the formation of the adaxial and the abaxial domains, respectively. While their importance is well documented, how their gradients help establish and maintain the adaxial-abaxial polarity is not clear. Scacchi, Paszkiewicz, Nguyen et al. show that tasiARF and miR165/6 are the at the center of a self-regulated and organ-autonomous genetic network that can be explained mathematically using a Turing diffusion model. Their computational effort is further supported by their complementary experimental data, which validated the novel genetic interactions predicted by their model. On top of explaining the self-sustained determination of leaf polarity, this model also accounts for the emergence of different leaf shapes within the same plant as well as between different species. Indeed, changing the values of the model’s parameters predicts the emergence of leaf morphologies other than the classic planar architecture. These findings are substantiated by experimental evidence in Arabidopsis mutants as well as similar evidence in other species. (Summary by Carlo Pasini @Crl_Psn) Nature Plants 10.1038/s41477-024-01634-x
The establishment of adaxial-abaxial polarity during the growth of the leaf primordium is a prerequisite for the formation of the flat, thin leaves observed in most (but not all) plants. The micro-RNAs tasiARF and miR165/6, which form concentration gradients within leaf primordia, are major contributors to the formation of the adaxial and the abaxial domains, respectively. While their importance is well documented, how their gradients help establish and maintain the adaxial-abaxial polarity is not clear. Scacchi, Paszkiewicz, Nguyen et al. show that tasiARF and miR165/6 are the at the center of a self-regulated and organ-autonomous genetic network that can be explained mathematically using a Turing diffusion model. Their computational effort is further supported by their complementary experimental data, which validated the novel genetic interactions predicted by their model. On top of explaining the self-sustained determination of leaf polarity, this model also accounts for the emergence of different leaf shapes within the same plant as well as between different species. Indeed, changing the values of the model’s parameters predicts the emergence of leaf morphologies other than the classic planar architecture. These findings are substantiated by experimental evidence in Arabidopsis mutants as well as similar evidence in other species. (Summary by Carlo Pasini @Crl_Psn) Nature Plants 10.1038/s41477-024-01634-x
Shade avoidance responses in Chinese white poplar rely on shared and unique roles of phytochrome-interacting factors
 Shade avoidance syndrome (SAS) is a set of adaptive growth responses to low red to far-red light ratios. SAS includes petiole and internode lengthening and upward bending of leaves (hyponasty). In Arabidopsis, PHYTOCHROME B (PHYB) and PHYTOCHROME INTERACTING FACTORS (PIFs) are implicated in these responses. Little is known, however, about the mechanism behind SAS in trees. Here, Sun et al. investigated how Populus tomentosa (Chinese white poplar) responds to simulated shade. Overexpression of PtoPHYB1/2 leads to attenuated SAS responses in shade-grown plants when compared to the wild-type, supporting previous reports that these photoreceptors play a conserved role in this species. Binding assays indicated that, as in Arabidopsis, PtoPIFs bind PtoPHYB1/2. Testing mutant and overexpressing lines of eight PtoPIF sequences revealed that while PtoPIF1/3.1/3.2 overexpression leads to taller and more shade-sensitive plants, those overexpressing PtoPIF8.1/8.2 are shorter than WT, yet hyponastic. It was also observed that, as in Arabidopsis, PtoPIFs induce the expression of the auxin biosynthesis gene PtoYUCCA8. These results show that while the main PHYB-PIF regulation module is conserved in P. tomentosa, PtoPIFs display both shared and unique functions in regulating SAS. (Summary by Johnatan Vilasboa @vilasjohn) Plant Cell Environ. 10.1111/pce.14853
Shade avoidance syndrome (SAS) is a set of adaptive growth responses to low red to far-red light ratios. SAS includes petiole and internode lengthening and upward bending of leaves (hyponasty). In Arabidopsis, PHYTOCHROME B (PHYB) and PHYTOCHROME INTERACTING FACTORS (PIFs) are implicated in these responses. Little is known, however, about the mechanism behind SAS in trees. Here, Sun et al. investigated how Populus tomentosa (Chinese white poplar) responds to simulated shade. Overexpression of PtoPHYB1/2 leads to attenuated SAS responses in shade-grown plants when compared to the wild-type, supporting previous reports that these photoreceptors play a conserved role in this species. Binding assays indicated that, as in Arabidopsis, PtoPIFs bind PtoPHYB1/2. Testing mutant and overexpressing lines of eight PtoPIF sequences revealed that while PtoPIF1/3.1/3.2 overexpression leads to taller and more shade-sensitive plants, those overexpressing PtoPIF8.1/8.2 are shorter than WT, yet hyponastic. It was also observed that, as in Arabidopsis, PtoPIFs induce the expression of the auxin biosynthesis gene PtoYUCCA8. These results show that while the main PHYB-PIF regulation module is conserved in P. tomentosa, PtoPIFs display both shared and unique functions in regulating SAS. (Summary by Johnatan Vilasboa @vilasjohn) Plant Cell Environ. 10.1111/pce.14853
Glutamate receptors shape plant systemic wound signaling and anti-herbivore defense
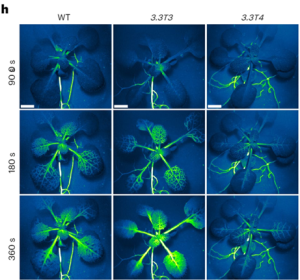 Systemic immunity, where local plant interactions with microbes or insects trigger enhanced resistance in distant organs, has parallels to the nervous system in animals. In plants, GLUTAMATE RECEPTOR-LIKE (GLR) genes have a pivotal role in long-distance wound signal transmission and the initiation of defense mechanisms including the accumulation of the defense hormone jasmonate (JA) at distal sites. Like the animal counterparts, plant GLRs function as ligand-gated Ca2+-permeable channels formed by homo- or hetero-tetrameric subunits. Despite this structural similarity, the regulatory components and molecular mechanisms governing plant GLR channel activity have remained elusive. In new work, Yan and colleagues unraveled the regulatory role of the cytoplasmic carboxy-terminal domain (CTD) in GLR3.3 function. Although GLR3.3 does not have a canonical calmodulin (CaM) binding domain, experimental studies showed that it does in fact bind CaM and that this CaM-binding domain in the CTD influences the desensitization of the GLR3.3 channel. Contrary to loss-of-function mutants compromised in defense, CRISPR-engineered GLR3.3 mutants with a five-amino-acid deletion in CTD were found to be a gain-of-function allele. This mutant exhibits prolonged slow wave potential and heightened JA-response, possibly due to decreased CaM-binding. Notably, this gain-of-function allele facilitated a more robust systemic JA response and improved anti-herbivore defense, all without impairing plant growth under normal conditions. This research offers insights into the intricate regulatory mechanisms of plant GLRs, paving the way for targeted strategies to fortify plant growth and defense. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-023-01578-8
Systemic immunity, where local plant interactions with microbes or insects trigger enhanced resistance in distant organs, has parallels to the nervous system in animals. In plants, GLUTAMATE RECEPTOR-LIKE (GLR) genes have a pivotal role in long-distance wound signal transmission and the initiation of defense mechanisms including the accumulation of the defense hormone jasmonate (JA) at distal sites. Like the animal counterparts, plant GLRs function as ligand-gated Ca2+-permeable channels formed by homo- or hetero-tetrameric subunits. Despite this structural similarity, the regulatory components and molecular mechanisms governing plant GLR channel activity have remained elusive. In new work, Yan and colleagues unraveled the regulatory role of the cytoplasmic carboxy-terminal domain (CTD) in GLR3.3 function. Although GLR3.3 does not have a canonical calmodulin (CaM) binding domain, experimental studies showed that it does in fact bind CaM and that this CaM-binding domain in the CTD influences the desensitization of the GLR3.3 channel. Contrary to loss-of-function mutants compromised in defense, CRISPR-engineered GLR3.3 mutants with a five-amino-acid deletion in CTD were found to be a gain-of-function allele. This mutant exhibits prolonged slow wave potential and heightened JA-response, possibly due to decreased CaM-binding. Notably, this gain-of-function allele facilitated a more robust systemic JA response and improved anti-herbivore defense, all without impairing plant growth under normal conditions. This research offers insights into the intricate regulatory mechanisms of plant GLRs, paving the way for targeted strategies to fortify plant growth and defense. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-023-01578-8
NAPstars: the new stars of the redox imaging toolbox!
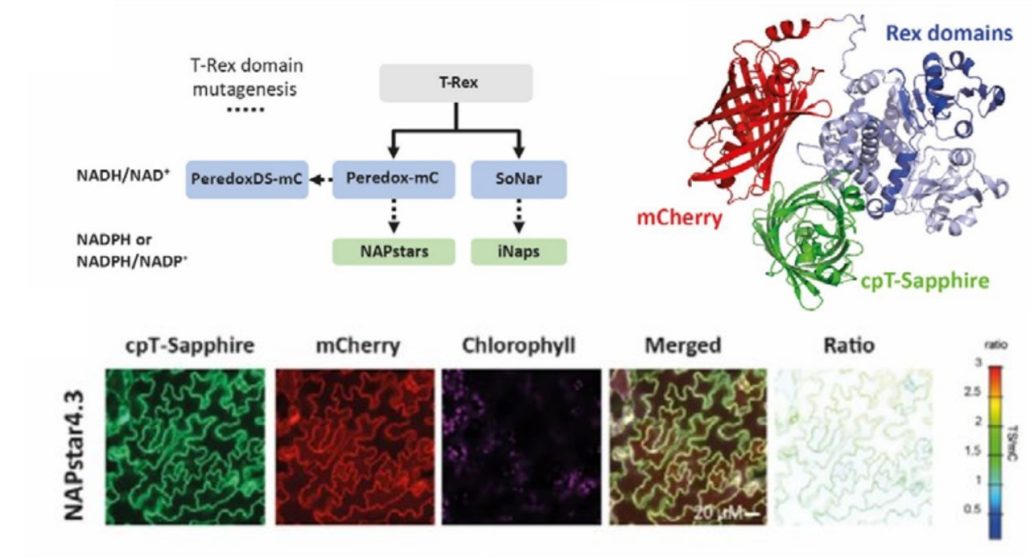 Redox reactions are an integral part of cellular metabolism and nicotinamide adenine dinucleotide phosphate or NADP is an essential component due to its capacity to be oxidized or reduced. To get a better understanding of cellular redox homeostasis, precise quantification and visualization of NADP redox states is vital. Despite the availability of biosensors for hydrogen peroxide, glutathione, and NAD redox state, accurate measurement of NADP redox state has been lacking, hindering detailed investigation of its role at the subcellular level. In a recent study, Scherschel et al. introduced NAPstars, the first family of genetically encoded fluorescent-based biosensors that can monitor the NADP redox state in cytosol and nucleus. Using these novel biosensors, they provide intriguing new insights into the regulation of the cytosolic NADP redox state in plants challenged with hypoxia, which are in stark contrast with the trend observed for NAD redox states, underscoring the need for specific tools to study redox biology. In contrast to what was previously thought, the glutathione system, which is closely connected with NADP redox state, emerged as the major reductive pathway in cells suffering oxidative stress. In conclusion, the introduction of NAPstars to the current toolbox of biosensors will allow improved accuracy in imaging and quantifying redox biology processes. Anticipating the development of variants targeting diverse subcellular compartments, these biosensors, in conjunction with other redox-related biosensors, promise a comprehensive understanding of redox dynamics in the entire cell. (Summary by Thomas Depaepe @thdpaepe) bioRxiv 10.1101/2024.02.14.580349
Redox reactions are an integral part of cellular metabolism and nicotinamide adenine dinucleotide phosphate or NADP is an essential component due to its capacity to be oxidized or reduced. To get a better understanding of cellular redox homeostasis, precise quantification and visualization of NADP redox states is vital. Despite the availability of biosensors for hydrogen peroxide, glutathione, and NAD redox state, accurate measurement of NADP redox state has been lacking, hindering detailed investigation of its role at the subcellular level. In a recent study, Scherschel et al. introduced NAPstars, the first family of genetically encoded fluorescent-based biosensors that can monitor the NADP redox state in cytosol and nucleus. Using these novel biosensors, they provide intriguing new insights into the regulation of the cytosolic NADP redox state in plants challenged with hypoxia, which are in stark contrast with the trend observed for NAD redox states, underscoring the need for specific tools to study redox biology. In contrast to what was previously thought, the glutathione system, which is closely connected with NADP redox state, emerged as the major reductive pathway in cells suffering oxidative stress. In conclusion, the introduction of NAPstars to the current toolbox of biosensors will allow improved accuracy in imaging and quantifying redox biology processes. Anticipating the development of variants targeting diverse subcellular compartments, these biosensors, in conjunction with other redox-related biosensors, promise a comprehensive understanding of redox dynamics in the entire cell. (Summary by Thomas Depaepe @thdpaepe) bioRxiv 10.1101/2024.02.14.580349
Visualizing plant intracellular inorganic orthophosphate distribution
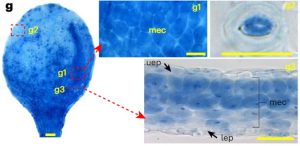 Phosphorus is the second most essential macronutrient in terms of limiting plant growth, acquired in the form of orthophosphate (Pi) by plant roots. The intricate processes of sensing, uptake, transport, storage, utilization, and cellular compartmentalization of Pi are finely orchestrated by a complex network of transporters and their regulators. The plant’s Pi status, encompassing both external and internal levels, propels a range of morphological and physiological adaptive responses collectively termed Pi Starvation Responses (PSRs). PSRs are indispensable not only for plant survival under Pi-starved conditions but also play a crucial role in symbiosis and plant immunity. This underscores the diverse functional significance of Pi signaling pathways. However, the accurate detection of Pi distribution at cellular and sub-cellular levels has proven to be a great challenge. Existing detection methods, using physical, biological, or chemical approaches, each possess inherent limitations, rendering direct visualization of Pi distribution in plant cells difficult. In a groundbreaking development, Guo and colleagues have introduced an efficient colorimetric staining method for Pi, based on the molybdenum blue assay. This methodology enables the observation of Pi distribution through microscopy at both cellular and subcellular levels, applicable to both living plant tissues and fixed sections. This breakthrough paves the way for new discoveries, shedding light on either novel components in Pi signaling or dissecting mechanistic insights into known regulators. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-023-01612-9
Phosphorus is the second most essential macronutrient in terms of limiting plant growth, acquired in the form of orthophosphate (Pi) by plant roots. The intricate processes of sensing, uptake, transport, storage, utilization, and cellular compartmentalization of Pi are finely orchestrated by a complex network of transporters and their regulators. The plant’s Pi status, encompassing both external and internal levels, propels a range of morphological and physiological adaptive responses collectively termed Pi Starvation Responses (PSRs). PSRs are indispensable not only for plant survival under Pi-starved conditions but also play a crucial role in symbiosis and plant immunity. This underscores the diverse functional significance of Pi signaling pathways. However, the accurate detection of Pi distribution at cellular and sub-cellular levels has proven to be a great challenge. Existing detection methods, using physical, biological, or chemical approaches, each possess inherent limitations, rendering direct visualization of Pi distribution in plant cells difficult. In a groundbreaking development, Guo and colleagues have introduced an efficient colorimetric staining method for Pi, based on the molybdenum blue assay. This methodology enables the observation of Pi distribution through microscopy at both cellular and subcellular levels, applicable to both living plant tissues and fixed sections. This breakthrough paves the way for new discoveries, shedding light on either novel components in Pi signaling or dissecting mechanistic insights into known regulators. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-023-01612-9
SIMPEL tool to elucidate dynamics of complex metabolism
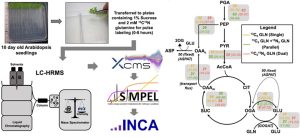 Cellular metabolism involves flux through metabolic routes, which can be measured using stable isotope (13C,15N) tracers. High-resolution mass spectrometry (HRMS) and transient isotope labeling studies offer an opportunity to get an in-depth understanding of metabolism in specific contexts. In this new work, Kambhampati et al. introduce Stable Isotope-assisted Metabolomics for Pathway Elucidation (SIMPEL) as a new tool that advances the analysis and interpretation of isotope-enriched HRMS datasets to elucidate operational metabolic pathways. SIMPEL is a data analysis tool that uses accurate mass information to group and compare isotope labeling and to cluster enriched metabolites based on their precursor-product relationships. It enables the detection and quantification of isotopically labeled metabolites in untargeted HRMS datasets for the exploration and characterization of new metabolic processes. The authors demonstrate SIMPEL’s effectiveness by two studies, a dual-isotope labeling flux experiment and a single-label lipidomic analysis. The SIMPEL tool is available in the R package and expands metabolomics investigations by including isotopologue signatures that are essential for quantifying metabolic flux. (Summary by Maneesh Lingwan @LingwanManeesh) Commun. Biol. 10.1038/s42003-024-05844-z
Cellular metabolism involves flux through metabolic routes, which can be measured using stable isotope (13C,15N) tracers. High-resolution mass spectrometry (HRMS) and transient isotope labeling studies offer an opportunity to get an in-depth understanding of metabolism in specific contexts. In this new work, Kambhampati et al. introduce Stable Isotope-assisted Metabolomics for Pathway Elucidation (SIMPEL) as a new tool that advances the analysis and interpretation of isotope-enriched HRMS datasets to elucidate operational metabolic pathways. SIMPEL is a data analysis tool that uses accurate mass information to group and compare isotope labeling and to cluster enriched metabolites based on their precursor-product relationships. It enables the detection and quantification of isotopically labeled metabolites in untargeted HRMS datasets for the exploration and characterization of new metabolic processes. The authors demonstrate SIMPEL’s effectiveness by two studies, a dual-isotope labeling flux experiment and a single-label lipidomic analysis. The SIMPEL tool is available in the R package and expands metabolomics investigations by including isotopologue signatures that are essential for quantifying metabolic flux. (Summary by Maneesh Lingwan @LingwanManeesh) Commun. Biol. 10.1038/s42003-024-05844-z