Plant Science Research Weekly: March 12, 2021
Review: Can smart nutrient applications optimize the plant’s hidden half to improve drought resistance?
 The root system has three key functions: support/anchoring, uptake of water, and uptake of nutrients. Many questions remain about how roots integrate all of these functions into the architecture of a single system. Here, Bardhan et al. explore whether modifying the nutrient environment (hence root’s foraging responses) can enhance root water uptake capabilities. The authors first describe the plastic (i.e., “moldable, changeable”) responses of roots to water deficit, and then the responses to localized nutrient distributions. The authors then look at the intersection of water/nutrient effects, and how these might be harnessed for improved drought tolerance. For example, a generally low availability of nutrients can promote greater root growth, and deep placement of fertilizers can promote deeper root growth, which can enhance water uptake. Spacing out nutrient application by time might also be employed to enhance water uptake. The available data for these approaches is sparce, so the key message of this article is to encourage further study, motivated by the vast yield losses that occur globally as a consequence of drought. (Summary by Mary Williams @PlantTeaching) Physiol. Plantarum 10.1111/ppl.13332
The root system has three key functions: support/anchoring, uptake of water, and uptake of nutrients. Many questions remain about how roots integrate all of these functions into the architecture of a single system. Here, Bardhan et al. explore whether modifying the nutrient environment (hence root’s foraging responses) can enhance root water uptake capabilities. The authors first describe the plastic (i.e., “moldable, changeable”) responses of roots to water deficit, and then the responses to localized nutrient distributions. The authors then look at the intersection of water/nutrient effects, and how these might be harnessed for improved drought tolerance. For example, a generally low availability of nutrients can promote greater root growth, and deep placement of fertilizers can promote deeper root growth, which can enhance water uptake. Spacing out nutrient application by time might also be employed to enhance water uptake. The available data for these approaches is sparce, so the key message of this article is to encourage further study, motivated by the vast yield losses that occur globally as a consequence of drought. (Summary by Mary Williams @PlantTeaching) Physiol. Plantarum 10.1111/ppl.13332
LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate
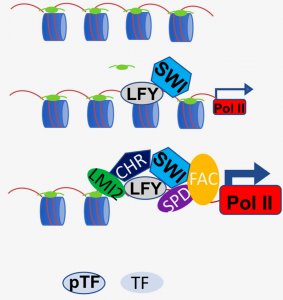 Master transcription factors (TFs) can activate specific genetic programs to reprogram cellular fate in the context of open chromatin. A special class of these proteins known as pioneer TFs are defined by their ability to trigger cell fate reprogramming by binding their cognate cis motifs in a nucleosome context both in vitro and in vivo, facilitating access of additional master TFs through chromatin opening. Pioneer TFs have been primarily studied in animals, however although pioneering activity has been proposed for several plant TFs it has not been established if any of these potential pioneering factors indeed contact nucleosome-occupied regulatory motifs. An interesting potential pioneering factor to investigate is LEAFY (LFY), an essential TF for floral fate that acts through upregulation of the floral commitment TF APETALA1 (AP1) and for which several lines of research hint at possible pioneer factor activity. A recent work by Jin et al. combined biochemical, structural and genomic approaches to unravel LFY mechanism of action and assess whether LFY indeed acts as a bona fide pioneer TF. The authors found that LFY can bind nucleosome-occupied AP1 target locus both in vitro and in vivo, consistent with what is observed for mammalian pioneer TFs. Furthermore, ChIP-qPCR and structural analyses revealed that LFY displays additional characteristic properties of mammalian pioneer TFs, such as chromatin opening through linker histone H1 displacement and subsequent chromatin remodeling as well as the ability to form very shallow DNA contacts, thus allowing for recruitment of additional factors to trigger cell reprogramming. This report calls attention to further identify, validate and characterize pioneer transcription factors in plants in order to better understand the basis of plant developmental plasticity and cell fate reprogramming. (Summary by Jesus Leon @jesussaur) Nature Comms 10.1038/s41467-020-20883-w
Master transcription factors (TFs) can activate specific genetic programs to reprogram cellular fate in the context of open chromatin. A special class of these proteins known as pioneer TFs are defined by their ability to trigger cell fate reprogramming by binding their cognate cis motifs in a nucleosome context both in vitro and in vivo, facilitating access of additional master TFs through chromatin opening. Pioneer TFs have been primarily studied in animals, however although pioneering activity has been proposed for several plant TFs it has not been established if any of these potential pioneering factors indeed contact nucleosome-occupied regulatory motifs. An interesting potential pioneering factor to investigate is LEAFY (LFY), an essential TF for floral fate that acts through upregulation of the floral commitment TF APETALA1 (AP1) and for which several lines of research hint at possible pioneer factor activity. A recent work by Jin et al. combined biochemical, structural and genomic approaches to unravel LFY mechanism of action and assess whether LFY indeed acts as a bona fide pioneer TF. The authors found that LFY can bind nucleosome-occupied AP1 target locus both in vitro and in vivo, consistent with what is observed for mammalian pioneer TFs. Furthermore, ChIP-qPCR and structural analyses revealed that LFY displays additional characteristic properties of mammalian pioneer TFs, such as chromatin opening through linker histone H1 displacement and subsequent chromatin remodeling as well as the ability to form very shallow DNA contacts, thus allowing for recruitment of additional factors to trigger cell reprogramming. This report calls attention to further identify, validate and characterize pioneer transcription factors in plants in order to better understand the basis of plant developmental plasticity and cell fate reprogramming. (Summary by Jesus Leon @jesussaur) Nature Comms 10.1038/s41467-020-20883-w
A complementary mechanism to the microRNA-mediated control of leaf size
 Plants form leaves with highly reproducible sizes and shapes by employing a conserved set of developmental regulators. During early leaf growth, a zone of high cell proliferation is formed towards the proximal end of the lamina. Proliferating cells progressively exit this zone and enter the distal half of the lamina where cell expansion and differentiation take place. GROWTH REGULATING FACTORs (GRFs) are key determinants of leaf size by promoting cell division in the proliferation zone. A non-coding microRNA, miR396 post-transcriptionally suppresses GRFs in the expansion zone and confines their activity to the proliferation zone. In their recent study, Beltramino et al. identified a set of GRFs present in virtually all members of Brassicaceae that are not targeted by miR396. Although they were known to perform functions in leaf development similar to the miR396-controlled GRFs, how their expression is spatially regulated in the leaf lamina has been unknown. The authors found that unlike miR396-regulated GRFs that undergo post-transcriptional regulation, the expression of GRF5, which is not targeted by miR396, is spatially constrained primarily through direct transcriptional suppression by AUXIN RESPONSE FACTOR2 (ARF2). In the absence of ARF2, the plant loses the control over the expression of GRF5, causing greater cell number and leaf size. Manipulating miR396-targeted GRFs together with the ARF2-GRF5 module resulted in additive changes on leaf size, hinting at a possibility of using this combination for agronomically desirable leaf traits. (Summary by Yadukrishnan Premachandran @yadukrishprem) Plant Physiol. 10.1093/plphys/kiab014
Plants form leaves with highly reproducible sizes and shapes by employing a conserved set of developmental regulators. During early leaf growth, a zone of high cell proliferation is formed towards the proximal end of the lamina. Proliferating cells progressively exit this zone and enter the distal half of the lamina where cell expansion and differentiation take place. GROWTH REGULATING FACTORs (GRFs) are key determinants of leaf size by promoting cell division in the proliferation zone. A non-coding microRNA, miR396 post-transcriptionally suppresses GRFs in the expansion zone and confines their activity to the proliferation zone. In their recent study, Beltramino et al. identified a set of GRFs present in virtually all members of Brassicaceae that are not targeted by miR396. Although they were known to perform functions in leaf development similar to the miR396-controlled GRFs, how their expression is spatially regulated in the leaf lamina has been unknown. The authors found that unlike miR396-regulated GRFs that undergo post-transcriptional regulation, the expression of GRF5, which is not targeted by miR396, is spatially constrained primarily through direct transcriptional suppression by AUXIN RESPONSE FACTOR2 (ARF2). In the absence of ARF2, the plant loses the control over the expression of GRF5, causing greater cell number and leaf size. Manipulating miR396-targeted GRFs together with the ARF2-GRF5 module resulted in additive changes on leaf size, hinting at a possibility of using this combination for agronomically desirable leaf traits. (Summary by Yadukrishnan Premachandran @yadukrishprem) Plant Physiol. 10.1093/plphys/kiab014
Host-interactor screens of Phytophthora infestans RXLR proteins reveal vesicle trafficking as a major effector-targeted process
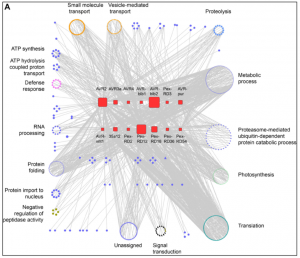 It’s like every spy movie you’ve ever seen. If you could infiltrate your opponent’s headquarters what would you target to disable your enemy? This is the question addressed in new work by Petre et al. More than ten years ago, a family of small proteins was identified, the RXLR proteins (named for a stretch of amino acids, Arg-X-Leu-Arg), that are effector proteins introduced from oomycete pathogens into host cells, allowing the pathogen to overrun the host cell. Here, the authors use an in planta co-immunoprecipitation “effectoromics” approach to investigate what host proteins these RXLR effectors target – an interactome. Besides an invaluable resource for further analysis, the study provides a clue into many of the host processes that a pathogen might target to gain the upper hand. Proteins that participate in vesicle trafficking form a large subset of these targets, supporting previous studies that show trafficking is essential for immune responses and defense (e.g., autophagy). (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koab069
It’s like every spy movie you’ve ever seen. If you could infiltrate your opponent’s headquarters what would you target to disable your enemy? This is the question addressed in new work by Petre et al. More than ten years ago, a family of small proteins was identified, the RXLR proteins (named for a stretch of amino acids, Arg-X-Leu-Arg), that are effector proteins introduced from oomycete pathogens into host cells, allowing the pathogen to overrun the host cell. Here, the authors use an in planta co-immunoprecipitation “effectoromics” approach to investigate what host proteins these RXLR effectors target – an interactome. Besides an invaluable resource for further analysis, the study provides a clue into many of the host processes that a pathogen might target to gain the upper hand. Proteins that participate in vesicle trafficking form a large subset of these targets, supporting previous studies that show trafficking is essential for immune responses and defense (e.g., autophagy). (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koab069
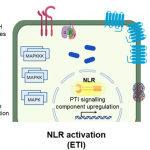
Not PTI or ETI: PTI and ETI
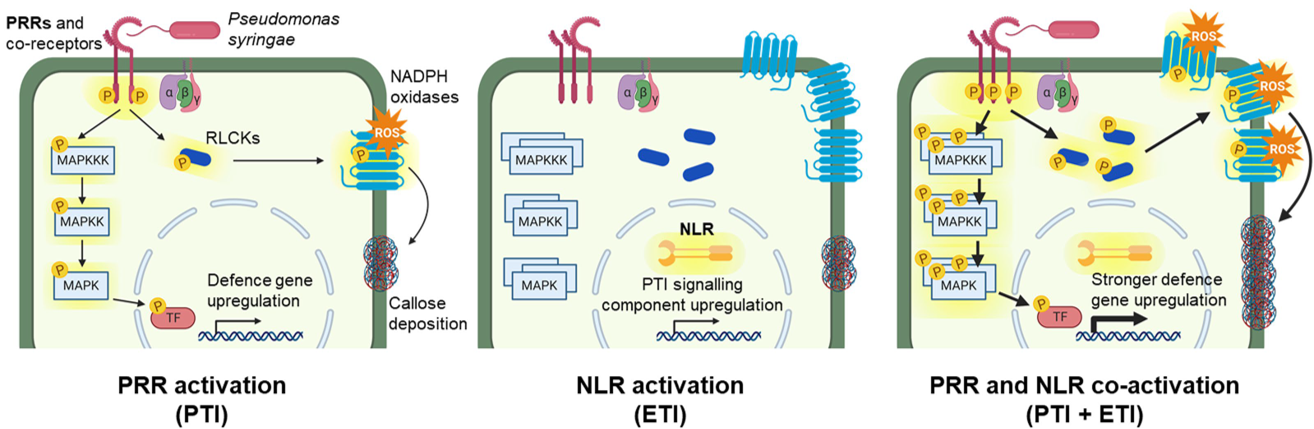 Two very exciting papers have come out this week in Nature that address a long-standing question about the relative contributions of two different plant immunity pathways. PTI (pathogen-triggered immunity) recognizes conserved pathogen signatures at the plant cell surface; a model system for PTI is the interaction between the bacteria flagellin protein (or the peptide flg22) and the cell-surface receptor FLS2. ETI (effector-triggered immunity) has been considered a second line of defense. Effectors are pathogen-derived proteins that are introduced into the plant cell, where they interfere with plant defenses. ETI is is the plant’s response to these effectors (or the consequences of the effectors), via NLRs (nucleotide-binding domain, leucine-rich-repeat containing receptors). These two papers, by Ngou et al. and by Yuan et al., ask what happens if ETI is triggered in the absence of PTI. In the first paper, the authors used an estradiol-inducible effector protein to trigger ETI, and in the second the authors used a genetic approach to knock-out the cell surface receptors and co-receptors. Both papers show clearly that neither PTI nor ETI confers immunity alone, but instead the whole is more than the sum of the parts, due to numerous positive interactions between the two systems (“mutual potentiations”). It’s a pleasure to see two complementary approaches that so clearly cement a new paradigm. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-021-03315-7 and 10.1038/s41586-021-03316-6
Two very exciting papers have come out this week in Nature that address a long-standing question about the relative contributions of two different plant immunity pathways. PTI (pathogen-triggered immunity) recognizes conserved pathogen signatures at the plant cell surface; a model system for PTI is the interaction between the bacteria flagellin protein (or the peptide flg22) and the cell-surface receptor FLS2. ETI (effector-triggered immunity) has been considered a second line of defense. Effectors are pathogen-derived proteins that are introduced into the plant cell, where they interfere with plant defenses. ETI is is the plant’s response to these effectors (or the consequences of the effectors), via NLRs (nucleotide-binding domain, leucine-rich-repeat containing receptors). These two papers, by Ngou et al. and by Yuan et al., ask what happens if ETI is triggered in the absence of PTI. In the first paper, the authors used an estradiol-inducible effector protein to trigger ETI, and in the second the authors used a genetic approach to knock-out the cell surface receptors and co-receptors. Both papers show clearly that neither PTI nor ETI confers immunity alone, but instead the whole is more than the sum of the parts, due to numerous positive interactions between the two systems (“mutual potentiations”). It’s a pleasure to see two complementary approaches that so clearly cement a new paradigm. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-021-03315-7 and 10.1038/s41586-021-03316-6