Plant Science Research Weekly: June 5th
Review: Functions of anionic lipids in plants
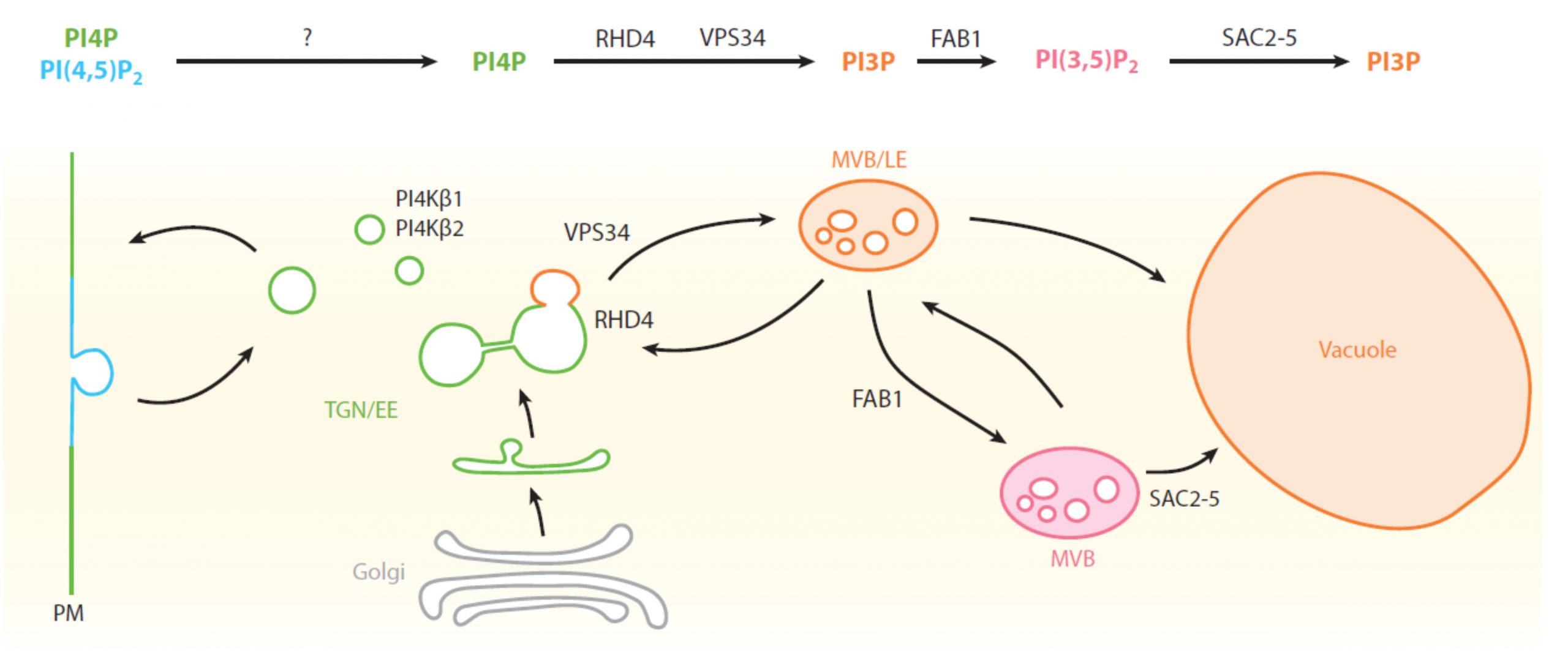 Moving materials within and out of cells requires that membranes carry identification labels, but when the membrane itself moves, that ID label must be updated. These requirements are met ingeniously by the anionic lipids, which are both a modifiable information system and simultaneously modify the physicochemical properties of the membranes in which they are embedded. This excellent review by Noack and Jaillais summarizes the functions of anionic lipids in plants, exploring the types of anionic lipids (e.g., phosphatidic acid, phosphatidylserine, phosphatidylinositol, and phosphatidylinositol phosphates), how they label compartments and serve as anchoring points for proteins, the enzymes that interconvert them, and their functions in processes as diverse as cell division, autophagy and arbuscular mycorrhizal symbiosis. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-081519-035910
Moving materials within and out of cells requires that membranes carry identification labels, but when the membrane itself moves, that ID label must be updated. These requirements are met ingeniously by the anionic lipids, which are both a modifiable information system and simultaneously modify the physicochemical properties of the membranes in which they are embedded. This excellent review by Noack and Jaillais summarizes the functions of anionic lipids in plants, exploring the types of anionic lipids (e.g., phosphatidic acid, phosphatidylserine, phosphatidylinositol, and phosphatidylinositol phosphates), how they label compartments and serve as anchoring points for proteins, the enzymes that interconvert them, and their functions in processes as diverse as cell division, autophagy and arbuscular mycorrhizal symbiosis. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-081519-035910
Review: Sequencing and analyzing the transcriptomes of a thousand species across the tree of life for green plants
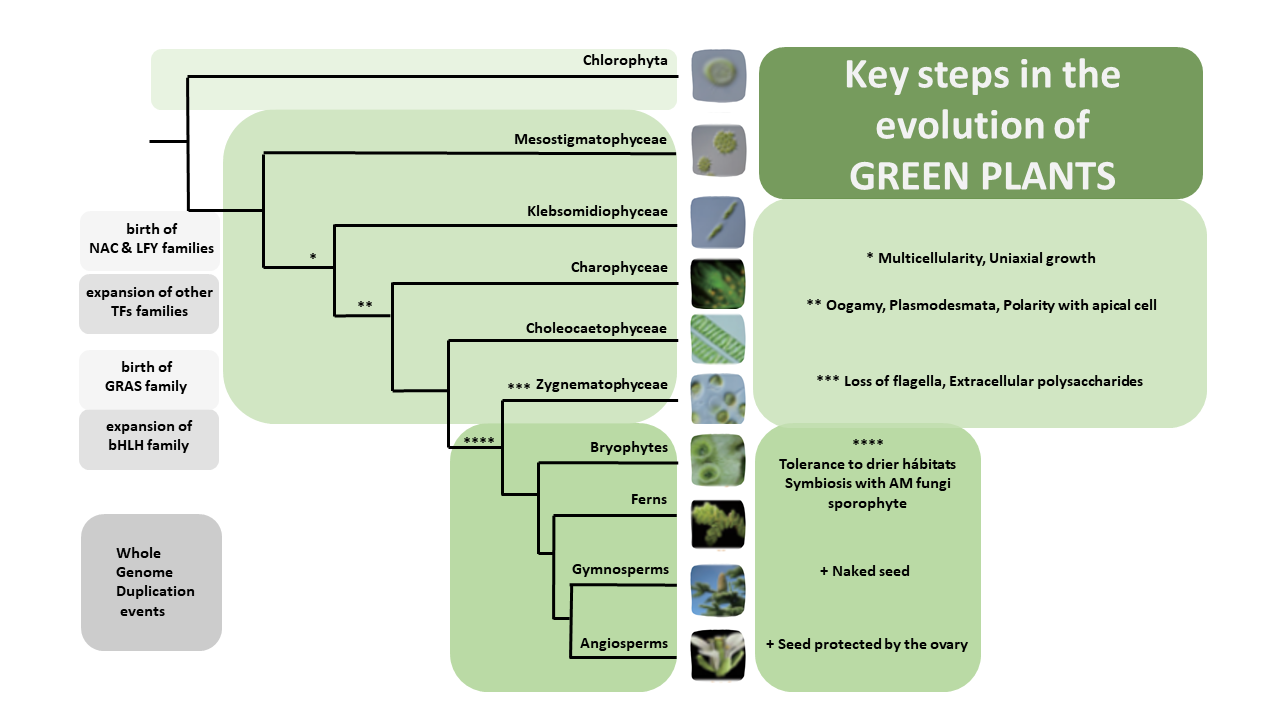 Over the past decade, Next Generation Sequencing (NGS) has been used for de novo assembly of crop genomes (i.e., tomato, potato) under the motto “If it tastes good, let’s sequence it”. By contrast, the One Thousand Plant (1KP) Initiative set out to obtain transcriptomic data of phylogenetically diverse green plant species without any apparent economic importance. Wong et al. review how the biodiversity in this large dataset is a valuable tool not only to address evolutionary questions but also to find original resources for genetic engineering. Phylotranscriptomic analysis of the 1KP dataset sheds new light on the diversification of green plant lineages, uncovering 138 novel Whole-Genome Duplication (WGD) events. Interestingly, paleopolyploidy was not found in the lycophyte Selaginella whereas at least five rounds of WGDs occurred in many angiosperms. Novel insights were gained on the origin and expansion of important multigene families encoding regulatory proteins. Surprisingly, regulatory genes thought to be specific to angiosperms, such as genetic determinants of flower development, were already present early in the evolution of streptophytes, the closest algal relatives of land plants. The 1KP dataset was also explored for biotechnological applications including studies of light-sensitive proteins to be employed in neuroscience and the elucidation of genetic networks underlying complex traits such as C4 photosynthesis. It will soon be feasible to sequence up to 10,000 genomes due to decreasing costs of NGS; these additional data will clarify discordances for critical nodes in the plant phylogenetic tree, and likely solve mysteries about the origin and evolution of green plants. (Summary and graphical abstract by Michela Osnato @michela_osnato) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042916-041040
Over the past decade, Next Generation Sequencing (NGS) has been used for de novo assembly of crop genomes (i.e., tomato, potato) under the motto “If it tastes good, let’s sequence it”. By contrast, the One Thousand Plant (1KP) Initiative set out to obtain transcriptomic data of phylogenetically diverse green plant species without any apparent economic importance. Wong et al. review how the biodiversity in this large dataset is a valuable tool not only to address evolutionary questions but also to find original resources for genetic engineering. Phylotranscriptomic analysis of the 1KP dataset sheds new light on the diversification of green plant lineages, uncovering 138 novel Whole-Genome Duplication (WGD) events. Interestingly, paleopolyploidy was not found in the lycophyte Selaginella whereas at least five rounds of WGDs occurred in many angiosperms. Novel insights were gained on the origin and expansion of important multigene families encoding regulatory proteins. Surprisingly, regulatory genes thought to be specific to angiosperms, such as genetic determinants of flower development, were already present early in the evolution of streptophytes, the closest algal relatives of land plants. The 1KP dataset was also explored for biotechnological applications including studies of light-sensitive proteins to be employed in neuroscience and the elucidation of genetic networks underlying complex traits such as C4 photosynthesis. It will soon be feasible to sequence up to 10,000 genomes due to decreasing costs of NGS; these additional data will clarify discordances for critical nodes in the plant phylogenetic tree, and likely solve mysteries about the origin and evolution of green plants. (Summary and graphical abstract by Michela Osnato @michela_osnato) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042916-041040
Multiple metabolic innovations and losses are associated with major transitions in land plant evolution
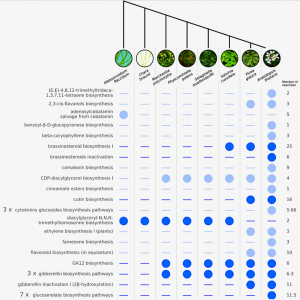 The colonization of land by a single streptophyte algae lineage around 450 million years ago culminated in the evolution and radiation of all terrestrial flora, the embryophytes. Adapting and thriving in the land environment required many morphological and physiological innovations, as well as the acquisition of novel metabolic pathways. However, metabolic pathway knowledge is biased towards angiosperm models. Cannell et al. explored metabolic gains or losses from streptophyte algae to angiosperms using a computational comparative approach: genome-scale metabolic pathway databases for eight representative species of major plant lineages were analyzed, supplemented by 64 and 305 additional plant genomes and transcriptomes. The analyses revealed that the complete repertoire for brassinosteroid biosynthesis/inactivation evolved in a stepwise fashion and is only fully present in seed plants, whereas most genes for gibberellin biosynthesis/inactivation are present in all land plants lineages, even in bryophytes, where gibberellins haven’t been experimentally detected. Moreover, the authors found that enzymes required for suberin and cutin production are found in angiosperms, while genes required for sporopollenin biosynthesis are conserved in all land plants. This paper identifies metabolic evolutionary trends across the plant phylogeny and opens a new research venue: the study of plant metabolism in extant non-angiosperm models. (Summary by Jesus Leon @jesussaur) Curr. Biol. 10.1016/j.cub.2020.02.086
The colonization of land by a single streptophyte algae lineage around 450 million years ago culminated in the evolution and radiation of all terrestrial flora, the embryophytes. Adapting and thriving in the land environment required many morphological and physiological innovations, as well as the acquisition of novel metabolic pathways. However, metabolic pathway knowledge is biased towards angiosperm models. Cannell et al. explored metabolic gains or losses from streptophyte algae to angiosperms using a computational comparative approach: genome-scale metabolic pathway databases for eight representative species of major plant lineages were analyzed, supplemented by 64 and 305 additional plant genomes and transcriptomes. The analyses revealed that the complete repertoire for brassinosteroid biosynthesis/inactivation evolved in a stepwise fashion and is only fully present in seed plants, whereas most genes for gibberellin biosynthesis/inactivation are present in all land plants lineages, even in bryophytes, where gibberellins haven’t been experimentally detected. Moreover, the authors found that enzymes required for suberin and cutin production are found in angiosperms, while genes required for sporopollenin biosynthesis are conserved in all land plants. This paper identifies metabolic evolutionary trends across the plant phylogeny and opens a new research venue: the study of plant metabolism in extant non-angiosperm models. (Summary by Jesus Leon @jesussaur) Curr. Biol. 10.1016/j.cub.2020.02.086
Genomic evidence for convergent evolution of gene clusters for momilactone
 Plants produce a rich diversity of chemical compounds. In fungi, the genes encoding specialized biosynthetic pathways are frequently arranged in contiguous loci forming biosynthetic gene clusters (BGC). In plants, BGCs are not common, however, some cases have been found in angiosperms. Among the known plant BGCs, a cluster responsible for the biosynthesis of the diterpenes momilactones -a broad range growth inhibitor-, was found in various grasses species but not in other plants. Interestingly, the moss Calohypnum plumiforme also produces momilactone. In this paper, Mao et al. sequenced its genome and found that the putative genes responsible for momilactone biosynthesis are also arranged in a cluster. They analyzed the co-expression and enzymatic activity of candidate genes using heterologous expression, confirming it is a BCG. Studying the evolution of this gene cluster in land plant species, they concluded that the arrangements of cluster genes are different in mosses than in grasses and might have evolved independently. This is the first report of a BGC in a non-seed plant, suggesting that they could be a more widespread genomic phenomenon that appeared early in the evolution of land plants. The genome of C. plumiforme will also help to understand bryophytes evolution. (Summary by Facundo Romani @facundoromani) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1914373117
Plants produce a rich diversity of chemical compounds. In fungi, the genes encoding specialized biosynthetic pathways are frequently arranged in contiguous loci forming biosynthetic gene clusters (BGC). In plants, BGCs are not common, however, some cases have been found in angiosperms. Among the known plant BGCs, a cluster responsible for the biosynthesis of the diterpenes momilactones -a broad range growth inhibitor-, was found in various grasses species but not in other plants. Interestingly, the moss Calohypnum plumiforme also produces momilactone. In this paper, Mao et al. sequenced its genome and found that the putative genes responsible for momilactone biosynthesis are also arranged in a cluster. They analyzed the co-expression and enzymatic activity of candidate genes using heterologous expression, confirming it is a BCG. Studying the evolution of this gene cluster in land plant species, they concluded that the arrangements of cluster genes are different in mosses than in grasses and might have evolved independently. This is the first report of a BGC in a non-seed plant, suggesting that they could be a more widespread genomic phenomenon that appeared early in the evolution of land plants. The genome of C. plumiforme will also help to understand bryophytes evolution. (Summary by Facundo Romani @facundoromani) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1914373117
Repurposed genes and the evolution of plant carnivory
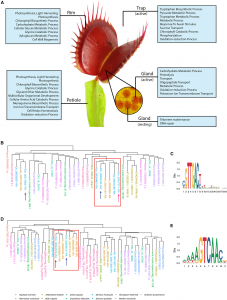 Carnivorous plants attract, trap, digest, and import nutrients from small animal prey, enabling these plants to thrive in nutrient-poor soil. Palfalvi et al. sequenced, annotated, and compared draft genomes from the family Droseraceae for the Venus flytrap (Dionaea muscipula), the waterwheel plant (Aldrovanda vesiculosa), and the sundew (Drosera spatulata). They identified an early whole-genome duplication that occurred at the base of the Droseraceae family that contributed to genetic diversification into novel functions for prey attraction, perception, digestion, and nutrient absorption. Genes were recruited for new carnivory-specific functions, such as from root nutrient transporters to leaf-derived trap nutrient absorption. Transcriptome analysis of ten tissues, including active and inactive traps, identified that nutrient transporters in the traps are only turned on after trap activation and that their promoters are associated with specific WRKY transcription factors that have been repurposed. These novel genomes show that the changing selective pressures during evolution of carnivory are associated with massive gene loss resulting in three of the gene-poorest plant genomes sequenced so far despite their physiological novelties. (Summary by Katy Dunning @plantmomkaty) Curr. Biol. 10.1016/j.cub.2020.04.051
Carnivorous plants attract, trap, digest, and import nutrients from small animal prey, enabling these plants to thrive in nutrient-poor soil. Palfalvi et al. sequenced, annotated, and compared draft genomes from the family Droseraceae for the Venus flytrap (Dionaea muscipula), the waterwheel plant (Aldrovanda vesiculosa), and the sundew (Drosera spatulata). They identified an early whole-genome duplication that occurred at the base of the Droseraceae family that contributed to genetic diversification into novel functions for prey attraction, perception, digestion, and nutrient absorption. Genes were recruited for new carnivory-specific functions, such as from root nutrient transporters to leaf-derived trap nutrient absorption. Transcriptome analysis of ten tissues, including active and inactive traps, identified that nutrient transporters in the traps are only turned on after trap activation and that their promoters are associated with specific WRKY transcription factors that have been repurposed. These novel genomes show that the changing selective pressures during evolution of carnivory are associated with massive gene loss resulting in three of the gene-poorest plant genomes sequenced so far despite their physiological novelties. (Summary by Katy Dunning @plantmomkaty) Curr. Biol. 10.1016/j.cub.2020.04.051
Dissecting the genome of star fruit (Averrhoa carambola L.)
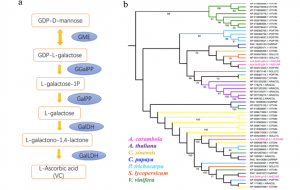 Starfruit is a sweet and sour fruit that has a shape of a five-point star. It belongs to the Oxalidaceae family and is a fruit of Averrhoa carambola, a species of tree native to tropical Southeast Asia. The fruit is consumed in many parts of the world and is known for its many economic, medicinal and nutritional benefits. Despite their popularity in a number of dishes, starfruit have not been well studied at the genetic level due to the lack of genome information. In this study, Fan et al. report the first draft of the genome information of the Oxalidaceae family. They assembled a total genome size of 470.51Mb and obtained 24,726 protein-coding genes including some nucleotide-binding site (NBS) genes that are important for disease resistance. Furthermore, the authors analyzed the evolutionary history of the fruit alongside 10 other species and found that A. carambola seperated from Populus trichocarpa (black cottonwood) approximately 94.5 million years ago. Finally, they identified some enzymes involved in the synthesis of nutritional metabolites including the vitamin C, vitamin B2, oxalate, and flavonoid metabolic pathways. This research provides an important resource for further studies on the nutritional, medicinal and economic importance of starfruit. (Summary by Modesta Abugu, @modestannedi) Hort. Res. 10.1038/s41438-020-0306-4
Starfruit is a sweet and sour fruit that has a shape of a five-point star. It belongs to the Oxalidaceae family and is a fruit of Averrhoa carambola, a species of tree native to tropical Southeast Asia. The fruit is consumed in many parts of the world and is known for its many economic, medicinal and nutritional benefits. Despite their popularity in a number of dishes, starfruit have not been well studied at the genetic level due to the lack of genome information. In this study, Fan et al. report the first draft of the genome information of the Oxalidaceae family. They assembled a total genome size of 470.51Mb and obtained 24,726 protein-coding genes including some nucleotide-binding site (NBS) genes that are important for disease resistance. Furthermore, the authors analyzed the evolutionary history of the fruit alongside 10 other species and found that A. carambola seperated from Populus trichocarpa (black cottonwood) approximately 94.5 million years ago. Finally, they identified some enzymes involved in the synthesis of nutritional metabolites including the vitamin C, vitamin B2, oxalate, and flavonoid metabolic pathways. This research provides an important resource for further studies on the nutritional, medicinal and economic importance of starfruit. (Summary by Modesta Abugu, @modestannedi) Hort. Res. 10.1038/s41438-020-0306-4
Improvement of predictive ability in maize hybrids by including dominance effects and marker x environment models ($)
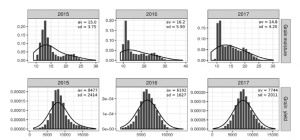 Heterosis is phenomenon that occurs when crossbred individuals show qualities (such as such as size, growth rate, fertility, and yield) that are superior to those of both parents. In maize, heterosis can be explained in terms of hybrid vigor, and this has been well studied using traditional breeding. However, testing multiple heterotic groups in different environments and breeding cycles is labor-intensive, presenting a bottleneck in maize breeding programs. The evolvement of genomic selection has made it possible to predict hybrid performance through evaluating the dominance gene action while accounting for other factors such as the impact of nonadditive effects and genotype x environment (G × E) interaction. In this study, Ferrão et al. used genomic selection to analyze the grain yield and moisture over three different cycles in multiple environments in maize. Their results showed the differences in performance for the two maize traits. For example, they demonstrated that including dominance effect in the model increased the predictive ability for grain yield by up to 30%, while additive models yielded better results for grain moisture. The authors also reported that including G × E interaction in the model increased accuracy in the prediction for one environment and thus, provides a premise for further studies on the prediction accuracy of G × E interaction models in multiple environments. (Summary by Modesta Abugu, @modestannedi) Crop Science: 10.1002/csc2.20096
Heterosis is phenomenon that occurs when crossbred individuals show qualities (such as such as size, growth rate, fertility, and yield) that are superior to those of both parents. In maize, heterosis can be explained in terms of hybrid vigor, and this has been well studied using traditional breeding. However, testing multiple heterotic groups in different environments and breeding cycles is labor-intensive, presenting a bottleneck in maize breeding programs. The evolvement of genomic selection has made it possible to predict hybrid performance through evaluating the dominance gene action while accounting for other factors such as the impact of nonadditive effects and genotype x environment (G × E) interaction. In this study, Ferrão et al. used genomic selection to analyze the grain yield and moisture over three different cycles in multiple environments in maize. Their results showed the differences in performance for the two maize traits. For example, they demonstrated that including dominance effect in the model increased the predictive ability for grain yield by up to 30%, while additive models yielded better results for grain moisture. The authors also reported that including G × E interaction in the model increased accuracy in the prediction for one environment and thus, provides a premise for further studies on the prediction accuracy of G × E interaction models in multiple environments. (Summary by Modesta Abugu, @modestannedi) Crop Science: 10.1002/csc2.20096
A gene knock-out that leads to seedless parthenocarpic fruits in Solanaceae plants ($)
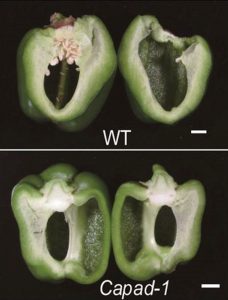 Parthenocarpy, or the ability to make fruit without fertilization, is desirable for many reasons including the opportunity to make seedless fruits and a greater resiliency in crop production in the face of climate change. Matsuo et al. identified a new gene involved in parthenocarpy, starting with a spontaneous parthenocarpic mutation arising in eggplant. The gene is named PARENTAL ADVICE-1 (Pad-1), from an old Japanese proverb that says that “Similar to eggplant flowers, none of the thousands of parental advices will be wasted.” Although fruit develops in the absence of pollination, seeds form when pad-1 mutants are pollinated, making these lines useful for breeding purposes. The Pad-1 gene encodes an aminotransferase with homology to the Arabidopsis VAS1 gene that has a role in auxin homeostasis, and pad-1 mutants show elevated auxin levels which may be why the ovaries develop. Knocking out the Pad-1 gene in tomato and pepper similarly produced seedless fruit, a trait that will prove popular to many cooks. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2001211117
Parthenocarpy, or the ability to make fruit without fertilization, is desirable for many reasons including the opportunity to make seedless fruits and a greater resiliency in crop production in the face of climate change. Matsuo et al. identified a new gene involved in parthenocarpy, starting with a spontaneous parthenocarpic mutation arising in eggplant. The gene is named PARENTAL ADVICE-1 (Pad-1), from an old Japanese proverb that says that “Similar to eggplant flowers, none of the thousands of parental advices will be wasted.” Although fruit develops in the absence of pollination, seeds form when pad-1 mutants are pollinated, making these lines useful for breeding purposes. The Pad-1 gene encodes an aminotransferase with homology to the Arabidopsis VAS1 gene that has a role in auxin homeostasis, and pad-1 mutants show elevated auxin levels which may be why the ovaries develop. Knocking out the Pad-1 gene in tomato and pepper similarly produced seedless fruit, a trait that will prove popular to many cooks. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2001211117
Water availability effects on germination, membrane stability and initial root growth of Agave lechuguilla and A. salmiana
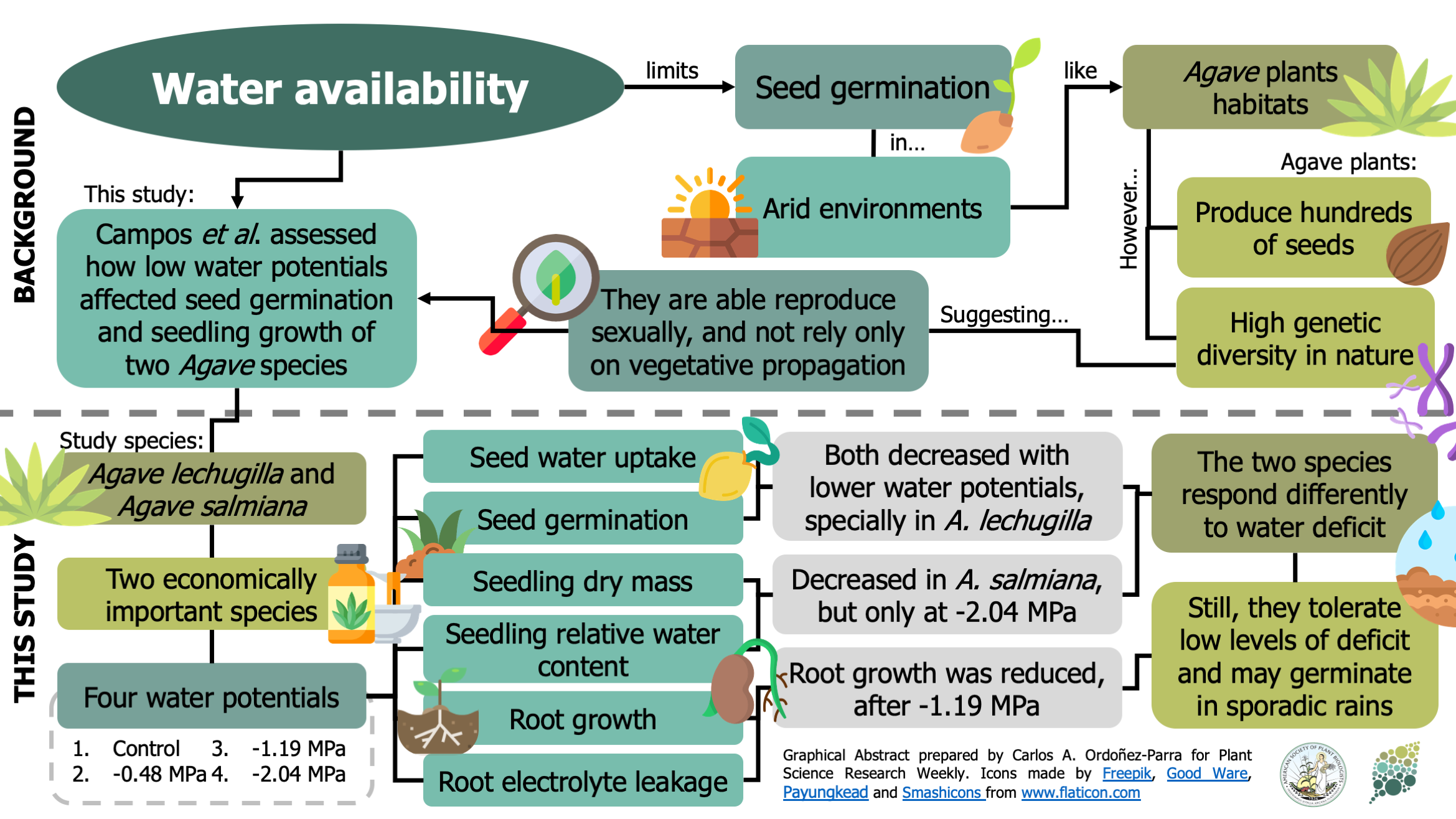 Low water availability limits seed germination in arid environments, such as the ones that Agave plants inhabit. As a result, vegetative propagation has been considered their most effective and successful method of reproduction. However, agaves produce several hundreds of seeds, and their natural populations show high genetic diversity, suggesting they do not rely solely on vegetative propagation. To test how water deficit limits sexual reproduction, Campos et al. assessed the impact of decreasing water potential on the germination and seedling growth of A. lechugilla and A. salmiana. Overall, lower water potentials reduced germination and root growth. However, these effects i) varied between species, ii) were significant only at the lowest water potentials (-1.19 and -2.04 MPa), and iii) did not inhibit germination completely. As a result, the authors show these species are adapted to germinate under variable water availability conditions. Moreover, since seedling dry mass was not affected by water potential, the authors suggest that seedlings could tolerate growing in these conditions, allowing the establishment of high genetic diversity populations via sexual reproduction. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Flora 10.1016/j.flora.2020.151606
Low water availability limits seed germination in arid environments, such as the ones that Agave plants inhabit. As a result, vegetative propagation has been considered their most effective and successful method of reproduction. However, agaves produce several hundreds of seeds, and their natural populations show high genetic diversity, suggesting they do not rely solely on vegetative propagation. To test how water deficit limits sexual reproduction, Campos et al. assessed the impact of decreasing water potential on the germination and seedling growth of A. lechugilla and A. salmiana. Overall, lower water potentials reduced germination and root growth. However, these effects i) varied between species, ii) were significant only at the lowest water potentials (-1.19 and -2.04 MPa), and iii) did not inhibit germination completely. As a result, the authors show these species are adapted to germinate under variable water availability conditions. Moreover, since seedling dry mass was not affected by water potential, the authors suggest that seedlings could tolerate growing in these conditions, allowing the establishment of high genetic diversity populations via sexual reproduction. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Flora 10.1016/j.flora.2020.151606
Post germination seedling establishment by ABA in response to light
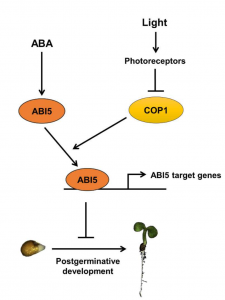 Abscisic acid (ABA), is implicated in reversible inhibition of seed germination (emergence of radicle) and post-germination seedling establishment (formation of green open cotyledons) during unfavorable conditions. Yadukrishnan et al. analyzed the role of light in ABA- mediated inhibition of post germination seedling growth and laid out a molecular framework using a combination of genetics and molecular biology. They showed that in dark, ABA mediated inhibition of post germination seedling establishment is enhanced and is dependent on the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) gene that encodes a E3 ligase. Further, they showed that in regulating the post germination seedling growth, COP1 acts downstream of ABSCISIC ACID INSENSITIVE5 (ABI5), an ABA responsive gene that encodes a basic leucine zipper transcription factor. At the protein level, COP1 helps ABI5 bind to the promoters of ABA responsive genes. Thus, in the dark, COP1 works with ABA to inhibit post-germination seedling establishment. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant J. 10.1111/tpj.14844
Abscisic acid (ABA), is implicated in reversible inhibition of seed germination (emergence of radicle) and post-germination seedling establishment (formation of green open cotyledons) during unfavorable conditions. Yadukrishnan et al. analyzed the role of light in ABA- mediated inhibition of post germination seedling growth and laid out a molecular framework using a combination of genetics and molecular biology. They showed that in dark, ABA mediated inhibition of post germination seedling establishment is enhanced and is dependent on the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) gene that encodes a E3 ligase. Further, they showed that in regulating the post germination seedling growth, COP1 acts downstream of ABSCISIC ACID INSENSITIVE5 (ABI5), an ABA responsive gene that encodes a basic leucine zipper transcription factor. At the protein level, COP1 helps ABI5 bind to the promoters of ABA responsive genes. Thus, in the dark, COP1 works with ABA to inhibit post-germination seedling establishment. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant J. 10.1111/tpj.14844
PIN FORMED 2 modulates the transport of arsenite in Arabidopsis thaliana
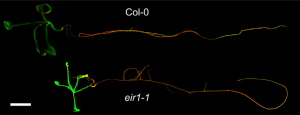 PIN FORMED (PIN) proteins are known for their directional auxin transport capacity. Ashraf et al. found that auxin transporter PINs have sequence similarity to bacterial arsenite transporter arsB, yeast Saccharomyces cerevisiae arsenite transporter SsAcr3, and arsenic hyperaccumulator fern Pteris vittata transporter PvAcr3, indicating that AtPIN2 is a possible arsenite efflux carrier. The authors found that arsenite specifically targets PIN2 protein trafficking and consequently alters auxin homeostasis. They additionally used radioactive arsenite and X-ray fluorescence imaging (XFI) coupled with X-ray absorption spectroscopy (XAS) to demonstrate in planta arsenite accumulation in the root tip in the absence of PIN2. Therefore, based on plant physiology, molecular and cellular biology, synchrotron imaging and radio-arsenite for transport assays, the authors demonstrated that the auxin transporter PIN2 also functions as an arsenite transporter. (Summary by Arif Ashraf) Plant Comms. 10.1016/j.xplc.2019.100009
PIN FORMED (PIN) proteins are known for their directional auxin transport capacity. Ashraf et al. found that auxin transporter PINs have sequence similarity to bacterial arsenite transporter arsB, yeast Saccharomyces cerevisiae arsenite transporter SsAcr3, and arsenic hyperaccumulator fern Pteris vittata transporter PvAcr3, indicating that AtPIN2 is a possible arsenite efflux carrier. The authors found that arsenite specifically targets PIN2 protein trafficking and consequently alters auxin homeostasis. They additionally used radioactive arsenite and X-ray fluorescence imaging (XFI) coupled with X-ray absorption spectroscopy (XAS) to demonstrate in planta arsenite accumulation in the root tip in the absence of PIN2. Therefore, based on plant physiology, molecular and cellular biology, synchrotron imaging and radio-arsenite for transport assays, the authors demonstrated that the auxin transporter PIN2 also functions as an arsenite transporter. (Summary by Arif Ashraf) Plant Comms. 10.1016/j.xplc.2019.100009
Measuring both microbial load and diversity with a single amplicon sequencing library
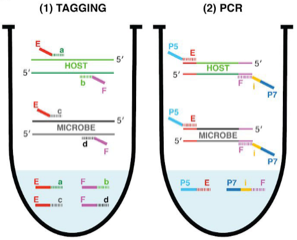 Amplicon sequencing of microbial DNA is a gold standard for analyzing the relative abundance of microbes in host-associated microbiomes. To gain more accurate insights into microbiome changes, it is crucial to know the absolute abundance of microbes, which can be analyzed by integrating relative abundance information with the total microbial load measured, which is measured by an additional experiment. However, such an approach is typically time consuming and noisy. To overcome this limitation, Lundberg et al. developed a simple and cost-effective strategy to measure both relative and absolute abundance of host-associated microbiomes from a single sequencing library. This approach, named host-associated microbe PCR (hamPCR), employs two steps of PCR reactions: the first limited-cycle PCR tags host and microbial target genes, then the second PCR amplifies all tagged products, followed by next-generation sequencing. PCR amplification with the common primers that target the tags can minimize amplification biases caused by differential primer efficiencies. The authors implemented a clever strategy to preadjust the host-to-microbe ratio in a sequencing library without losing information, which helps improve accuracy and cost efficiency in analyzing samples dominated by the host or microbial DNA. The method performed well for three amplicons (plants, bacteria, and oomycetes) and is applicable to a crop with a large genome and a non-plant host (nematode worms). In summary, hamPCR is a robust and easy-to-implement method that offers an accurate picture of the host-associated microbiome. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.19.103937
Amplicon sequencing of microbial DNA is a gold standard for analyzing the relative abundance of microbes in host-associated microbiomes. To gain more accurate insights into microbiome changes, it is crucial to know the absolute abundance of microbes, which can be analyzed by integrating relative abundance information with the total microbial load measured, which is measured by an additional experiment. However, such an approach is typically time consuming and noisy. To overcome this limitation, Lundberg et al. developed a simple and cost-effective strategy to measure both relative and absolute abundance of host-associated microbiomes from a single sequencing library. This approach, named host-associated microbe PCR (hamPCR), employs two steps of PCR reactions: the first limited-cycle PCR tags host and microbial target genes, then the second PCR amplifies all tagged products, followed by next-generation sequencing. PCR amplification with the common primers that target the tags can minimize amplification biases caused by differential primer efficiencies. The authors implemented a clever strategy to preadjust the host-to-microbe ratio in a sequencing library without losing information, which helps improve accuracy and cost efficiency in analyzing samples dominated by the host or microbial DNA. The method performed well for three amplicons (plants, bacteria, and oomycetes) and is applicable to a crop with a large genome and a non-plant host (nematode worms). In summary, hamPCR is a robust and easy-to-implement method that offers an accurate picture of the host-associated microbiome. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.19.103937
A chemical elicitor, 4- fluorophenoxyacetic acid suppresses insect pest populations and increases crop yields
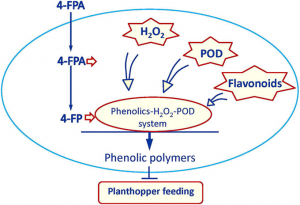 Plant strengtheners, synthetic chemical elicitors, have been shown to enhance plant resistance against various insect pests without toxic effects on the environment, but evidence is lacking for a significant increase in crop growth and yield after using these elicitors. To address this, Wang et al. studied 4-fluorophenoxyacetic acid (4-FPA), which in rice fields infested with white-backed planthopper led to a reduction in the insects’ survival rate. Treatment with 4-FPA induces production of peroxidases, hydrogen peroxide, and flavonoids. Additionally, deposition of flavonoid polymers in the parenchyma cells of the leaf sheath inhibits the plant hopper’s stylet from reaching the phloem sap. Greenhouse and field tests on growth and yield of rice after 4-FDA treatment showed growth recovery and significantly higher yield compared to control plants. Studies of 4-FDA in other cereals showed broad-spectrum resistance to piercing-sucking insect pests. In conclusion, this study showed 4-FDA as a promising chemical elicitor in controlling piercing and sucking insect pests. (Summary by Sunita Pathak @psunita980 ) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2003742117
Plant strengtheners, synthetic chemical elicitors, have been shown to enhance plant resistance against various insect pests without toxic effects on the environment, but evidence is lacking for a significant increase in crop growth and yield after using these elicitors. To address this, Wang et al. studied 4-fluorophenoxyacetic acid (4-FPA), which in rice fields infested with white-backed planthopper led to a reduction in the insects’ survival rate. Treatment with 4-FPA induces production of peroxidases, hydrogen peroxide, and flavonoids. Additionally, deposition of flavonoid polymers in the parenchyma cells of the leaf sheath inhibits the plant hopper’s stylet from reaching the phloem sap. Greenhouse and field tests on growth and yield of rice after 4-FDA treatment showed growth recovery and significantly higher yield compared to control plants. Studies of 4-FDA in other cereals showed broad-spectrum resistance to piercing-sucking insect pests. In conclusion, this study showed 4-FDA as a promising chemical elicitor in controlling piercing and sucking insect pests. (Summary by Sunita Pathak @psunita980 ) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2003742117
STRESS INDUCED FACTOR 2 regulates arabidopsis stomatal immunity through phosphorylation of the anion channel SLAC1 ($)
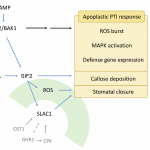
 Plants detect microbes by pattern recognition receptors (PRRs) that sense common conserved structures called microbe-associated molecular patterns (MAMPs) and trigger the innate immunity responses. Chan et al. identified a new player in the Arabidopsis immune response known as STRESS INDUCED FACTOR 2 (SIF2) and demonstrated that the knock-out mutant of SIF2 is more susceptible to bacteria. Upon flg22 or ABA treatment, sif2 shows impaired stomatal closure. When overexpressed, SIF2 protein increases resistance to bacteria and leads to constitutive stomatal closure. SIF2 interacts with other PRRs including FLS2 and the co-receptor BAK1, as wells as the guard cell anion channel SLAC1, a key factor in the control of osmotic pressure in guard cells. SIF2 contains a cytoplasmic kinase domain that phosphorylates SLAC1, activating its ion channel activity and promoting stomatal closure. Thus, SIF2 has a role in stomtal immunity. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.19.00578
Plants detect microbes by pattern recognition receptors (PRRs) that sense common conserved structures called microbe-associated molecular patterns (MAMPs) and trigger the innate immunity responses. Chan et al. identified a new player in the Arabidopsis immune response known as STRESS INDUCED FACTOR 2 (SIF2) and demonstrated that the knock-out mutant of SIF2 is more susceptible to bacteria. Upon flg22 or ABA treatment, sif2 shows impaired stomatal closure. When overexpressed, SIF2 protein increases resistance to bacteria and leads to constitutive stomatal closure. SIF2 interacts with other PRRs including FLS2 and the co-receptor BAK1, as wells as the guard cell anion channel SLAC1, a key factor in the control of osmotic pressure in guard cells. SIF2 contains a cytoplasmic kinase domain that phosphorylates SLAC1, activating its ion channel activity and promoting stomatal closure. Thus, SIF2 has a role in stomtal immunity. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.19.00578