Plant Science Research Weekly: June 21st
Guest editor: Magdalena Julkowska
Magda is a PostDoc at King Abdullah University for Science and Technology (KAUST, Saudi Arabia) working with Prof. Mark Tester. Her main interests are (1) salt-induced changes in the root-to-shoot ratio in Arabidopsis, (2) study the expression patterns in plants with enhanced sodium accumulation in their roots and (3) development of tools for data analysis based on R/Shiny. She is passionate about capturing plant architecture using simple models, understanding plant physiology and salinity tolerance.
A Transcription Factor, OsMADS57, Regulates Long-Distance Nitrate Transport and Root Elongation (Plant Phys.)
 Nitrogen is a critical macronutrient for plant growth and reproduction, and it’s largely present as ammonium in flooded and acidic soils. However, structurally porous tissue in rice roots, called aerenchyma, transfer oxygen to the root system in flooded paddies, increasing nitrate abundance. Huang et al. used a transcription factor, OsMADS57, to study the complex regulation of nitrogen homeostasis in a hydroponic setting. The group used mutants and overexpressing rice lines of OsMADS57, along with nitrogen isotope labeling to demonstrate that MADS57 regulates the root to shoot transport of nitrate. This was confirmed through RT-qPCR analysis, which showed a MADS57 transcriptional effect on members of the OsNRT2 family and nitrate reductase. Furthermore, a yeast 1-hybrid assay and transient tobacco expression revealed that MADS57 binds a specific CArG box in the promoter of NRT2.3a. Lastly, OsMADS57 was shown to alter auxin accumulation and growth of seminal rice roots. Taken together, these results highlight an additional regulator of nitrogen homeostasis and root growth in rice. (Summary by Nathan Scinto-Madonich) Plant Phys. 10.1104/pp.19.00142
Nitrogen is a critical macronutrient for plant growth and reproduction, and it’s largely present as ammonium in flooded and acidic soils. However, structurally porous tissue in rice roots, called aerenchyma, transfer oxygen to the root system in flooded paddies, increasing nitrate abundance. Huang et al. used a transcription factor, OsMADS57, to study the complex regulation of nitrogen homeostasis in a hydroponic setting. The group used mutants and overexpressing rice lines of OsMADS57, along with nitrogen isotope labeling to demonstrate that MADS57 regulates the root to shoot transport of nitrate. This was confirmed through RT-qPCR analysis, which showed a MADS57 transcriptional effect on members of the OsNRT2 family and nitrate reductase. Furthermore, a yeast 1-hybrid assay and transient tobacco expression revealed that MADS57 binds a specific CArG box in the promoter of NRT2.3a. Lastly, OsMADS57 was shown to alter auxin accumulation and growth of seminal rice roots. Taken together, these results highlight an additional regulator of nitrogen homeostasis and root growth in rice. (Summary by Nathan Scinto-Madonich) Plant Phys. 10.1104/pp.19.00142
Breeding improves wheat productivity under contrasting agrochemical input levels (Nature Biotech.)
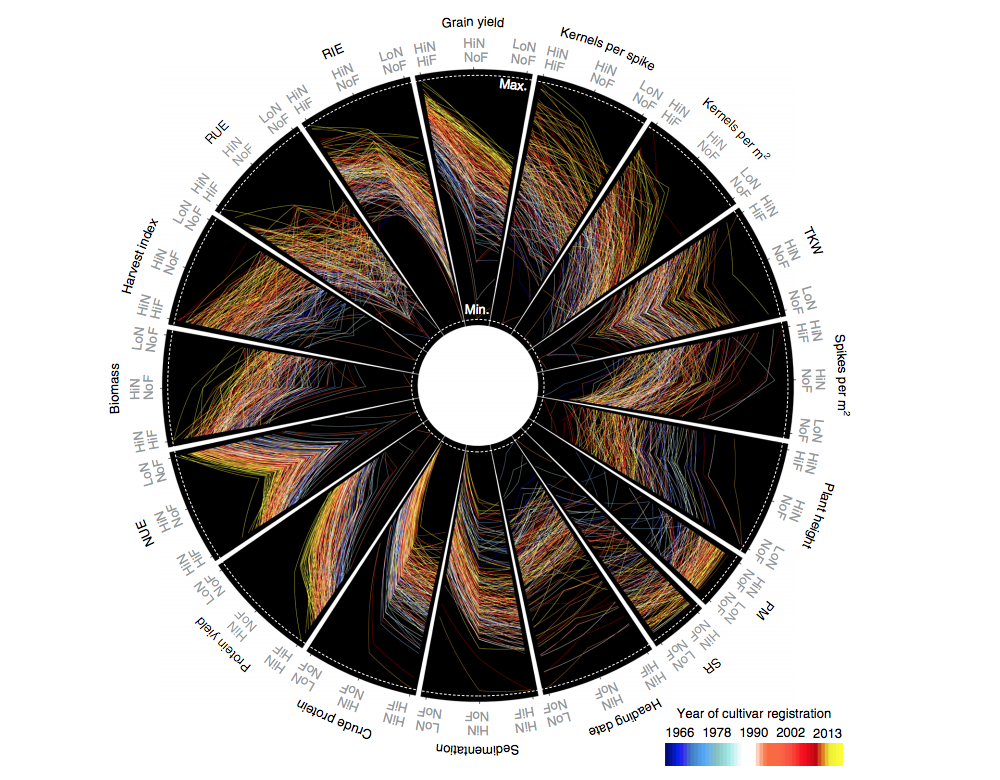 Wheat breeding programs are releasing new cultivars almost every year for improved yield potential. But is this intensive breeding not compromising the plant performance under adverse environmental conditions? Voss-Fels et al. studied the elite cultivars of winter wheat released during the last 50 years, across multiple field trials with and without application of fertilizer, best-practice fungicide, and mild drought stress. The cultivars released most recently were performing best in both optimal and sub-optimal conditions. Interestingly, no evidence was found for a long-term reduction in genetic diversity as a result of breeding. By developing a new haplotype-based approach genomic prediction method, Voss-Fels and colleagues were able to identify genomic loci contributing to the yield in the population of elite wheat varieties, which could further increase the yield potential up to 23% compared to the current best-performing varieties. (Summarized by Magdalena Julkowska) Nature Biotech. 10.1038/s41477-019-0445-5
Wheat breeding programs are releasing new cultivars almost every year for improved yield potential. But is this intensive breeding not compromising the plant performance under adverse environmental conditions? Voss-Fels et al. studied the elite cultivars of winter wheat released during the last 50 years, across multiple field trials with and without application of fertilizer, best-practice fungicide, and mild drought stress. The cultivars released most recently were performing best in both optimal and sub-optimal conditions. Interestingly, no evidence was found for a long-term reduction in genetic diversity as a result of breeding. By developing a new haplotype-based approach genomic prediction method, Voss-Fels and colleagues were able to identify genomic loci contributing to the yield in the population of elite wheat varieties, which could further increase the yield potential up to 23% compared to the current best-performing varieties. (Summarized by Magdalena Julkowska) Nature Biotech. 10.1038/s41477-019-0445-5
A specialized metabolic network selectively modulates Arabidopsis root microbiota (Science)
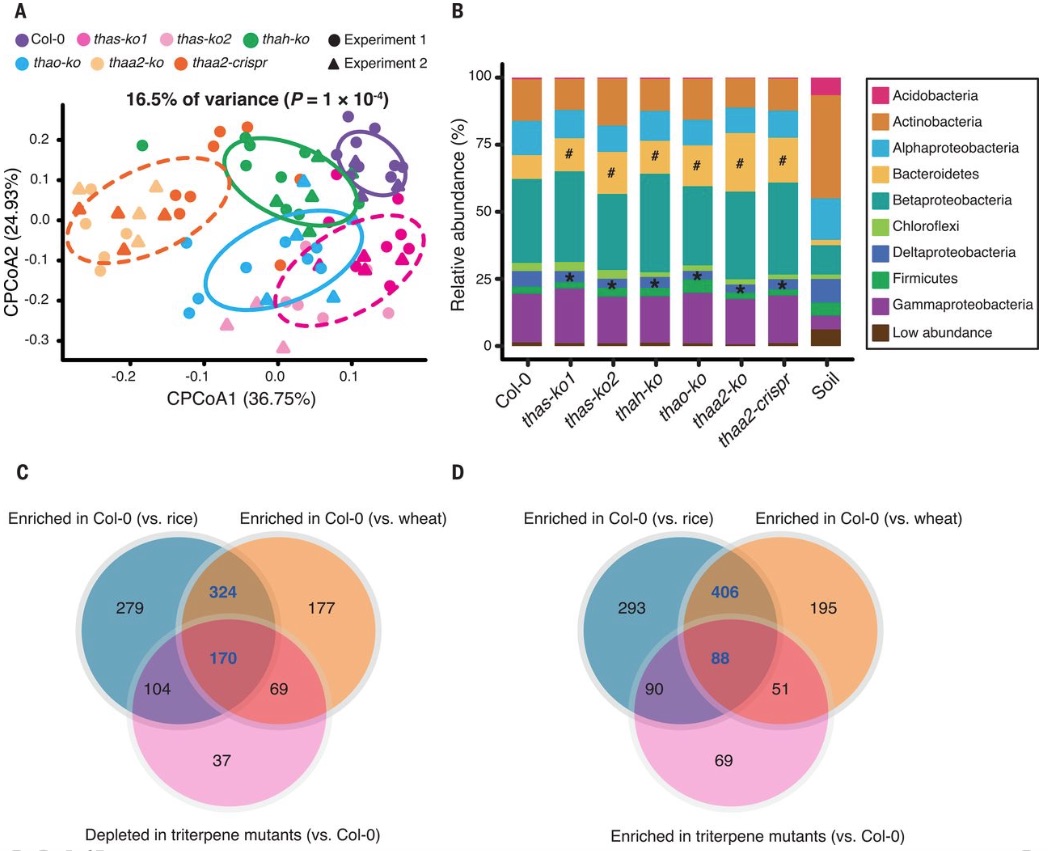
Plants have evolved to adapt to varied environmental niches. The ability to produce specialized metabolites is one of the evolved characteristics which help plants to thrive through varied biotic and abiotic conditions. Plant-associated microbes are always known to have a drastic impact on plant growth and fitness. Although, the knowledge about the factors responsible for defining microbiome by the plant is elusive. In a recently published research article by Huang et al., has shown the plant produced specialized metabolites such as triterpenes has a strong effect on deciding the root associated microbiome. Triterpenes are specifically known for their role in plant defense and signaling. In the present research article, they have identified and filled the gaps in biosynthesis pathways of different types of triterpenes, derived from thalianol and arabidiol gene clusters. Genes such as THAS, THAH, THAO, and THAA2 are involved in multiple pathways. The mutant lines of these genes showed to assemble different root microbiota as compared to the wild type. The disruption in the biosynthesis of triterpene metabolites cause the shifts in root-associated microbial community structure at taxa level. The authors have demonstrated by using the isolated microbial members, that the selective metabolites are used as a carbon source for growth. (Summarized by Mugdha Sabale) Science 10.1126/science.aau6389
A chloroplast-localized mitochondrial calcium uniporter transduces osmotic stress in Arabidopsis (Nature Plants)
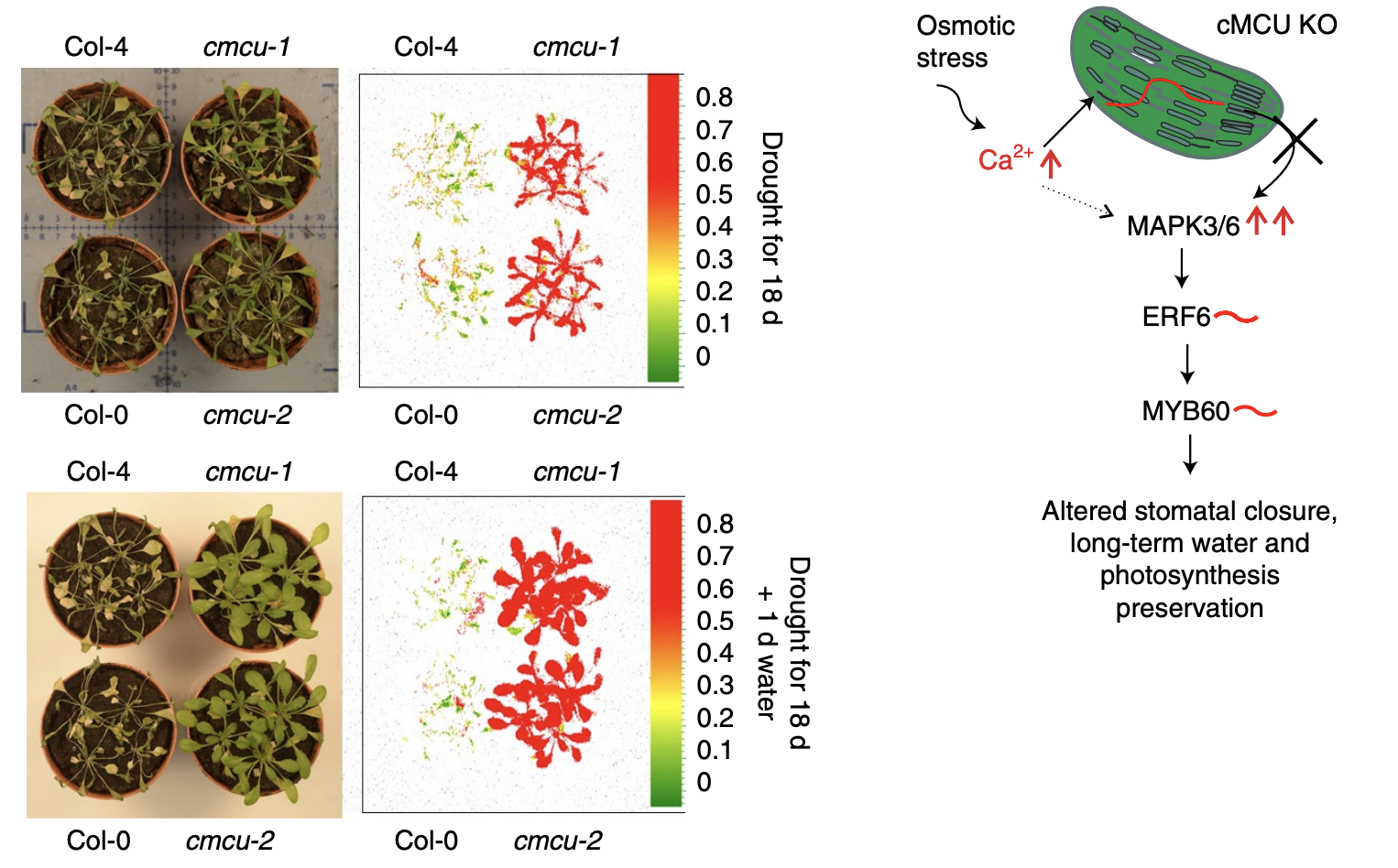 Calcium and chloroplasts are both at the heart of the signal transduction during environmental stress. Teardo et al. studied whether the chloroplast-localized mitochondrial calcium uniporter has a role in stress signaling. The group identified 6 homologs of mitochondrial calcium uniporter (MCU) in Arabidopsis, but only one (cMCU) was carrying a clear chloroplast localization signature. The cMCU localized to the outer chloroplast envelope in vivo where it mediated uptake of calcium ions. The mutants of cMCU showed extended calcium waves in response to osmotic stress and reduced calcium levels in the stroma. Interestingly, the effect of cMCU was detected only when plants were exposed to light, but not during the dark period. The mutants of cMCU had reduced stomatal aperture and were able to better maintain their photosynthetic efficiency during drought and recovery period. This resulted in increased tolerance to drought. Work by Teadro et al. shows how small changes in calcium signaling can result in significant alterations of environmental resilience. (Summarized by Magdalena Julkowska) Nature Plants 10.1038/s41477-019-0434-8
Calcium and chloroplasts are both at the heart of the signal transduction during environmental stress. Teardo et al. studied whether the chloroplast-localized mitochondrial calcium uniporter has a role in stress signaling. The group identified 6 homologs of mitochondrial calcium uniporter (MCU) in Arabidopsis, but only one (cMCU) was carrying a clear chloroplast localization signature. The cMCU localized to the outer chloroplast envelope in vivo where it mediated uptake of calcium ions. The mutants of cMCU showed extended calcium waves in response to osmotic stress and reduced calcium levels in the stroma. Interestingly, the effect of cMCU was detected only when plants were exposed to light, but not during the dark period. The mutants of cMCU had reduced stomatal aperture and were able to better maintain their photosynthetic efficiency during drought and recovery period. This resulted in increased tolerance to drought. Work by Teadro et al. shows how small changes in calcium signaling can result in significant alterations of environmental resilience. (Summarized by Magdalena Julkowska) Nature Plants 10.1038/s41477-019-0434-8
QTL × environment interactions underlie adaptive divergence in switchgrass across a large latitudinal gradient (PNAS)
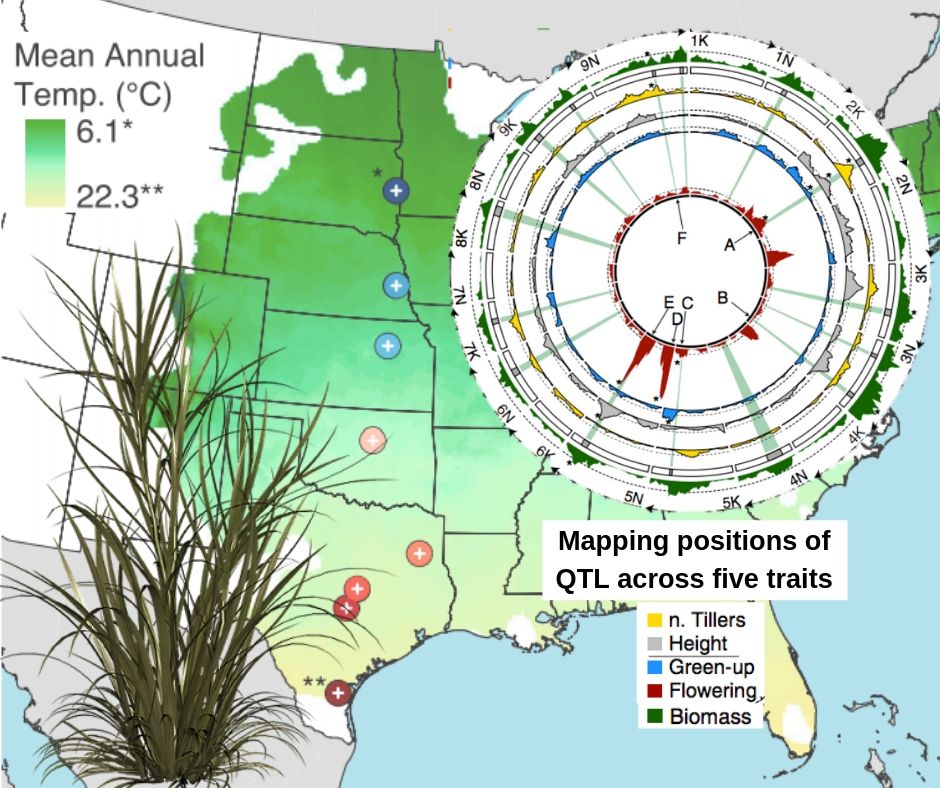
The main problem to elucidate the genetic basis in regards to a plant local adaptation is the geographic and climatic biased scope of the tests. Here, Lowry et al. overcame this by evaluating the contributions of individual genetic loci to the adaptation of outcrossing switchgrass (Panicum virgatum L.) across 10 geographic regions at the central United States. Highly divergent lowland and upland ecotypes were used to develop a four-way outbred mapping population, where the crosses design was key to quantify the differences in effects of the alleles. The authors mapped multiple significant Quantitative Trait Loci (QTL) for five traits: biomass, flowering and green-up time, plant height and tiller number. Most of the QTL showed an important genotype-by-environment (G×E) effect. However, the results suggested that the same loci do not consistently contribute to the divergence between the ecotypes. Furthermore, a two-way comparison of the biomass QTL additive effect looking for trade-offs showed they were weak, rare or undetectable, which implies a gene flow restriction between ecotypes, allowing to breed lines that perform well across large areas. (Summary by Ana Valladares) PNAS 10.1073/pnas.1821543116
Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading (Nature Plants)
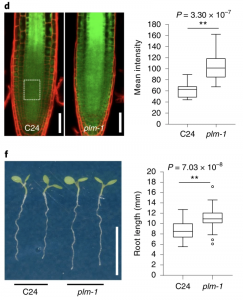 Regulated transport of molecules from source to sink is essential for providing necessary molecules including photoassimilates to the required tissue. This mechanism is crucial for development and plant growth in response to stimuli and it occurs through plasmodesmata. In a previous report, CALLOSE SYNTHASE 3 (CALS3) was identified to be necessary for the permeability of molecules via plasmodesmata and root development. In this paper Yan et al., have identified, PHLOEM UNLOADING MODULATOR (PLM) through a genetic screen for suppression of cals3-1d gain of function mutant which has an enhanced phloem unloading. PLM has been identified to be involved in the sphingolipid metabolism and the loss of function mutant, plm has altered endoplasmic reticulum-plasma membrane tethering which causes a change in the plasmodesmata structure. This change in sphingolipid metabolism could have caused the elevated unloading at the phloem pole pericycle –endodermis interface. Thus one more regulator, PLM involved in phloem unloading and its function in plasmodesmata mediated molecular trafficking has been identified. (Summarized by Suresh Damodaran) Nature Plants 10.1038/s41477-019-0429-5
Regulated transport of molecules from source to sink is essential for providing necessary molecules including photoassimilates to the required tissue. This mechanism is crucial for development and plant growth in response to stimuli and it occurs through plasmodesmata. In a previous report, CALLOSE SYNTHASE 3 (CALS3) was identified to be necessary for the permeability of molecules via plasmodesmata and root development. In this paper Yan et al., have identified, PHLOEM UNLOADING MODULATOR (PLM) through a genetic screen for suppression of cals3-1d gain of function mutant which has an enhanced phloem unloading. PLM has been identified to be involved in the sphingolipid metabolism and the loss of function mutant, plm has altered endoplasmic reticulum-plasma membrane tethering which causes a change in the plasmodesmata structure. This change in sphingolipid metabolism could have caused the elevated unloading at the phloem pole pericycle –endodermis interface. Thus one more regulator, PLM involved in phloem unloading and its function in plasmodesmata mediated molecular trafficking has been identified. (Summarized by Suresh Damodaran) Nature Plants 10.1038/s41477-019-0429-5
Paleogenomic insights into the origins of French grapevine diversity (Nature Plants)
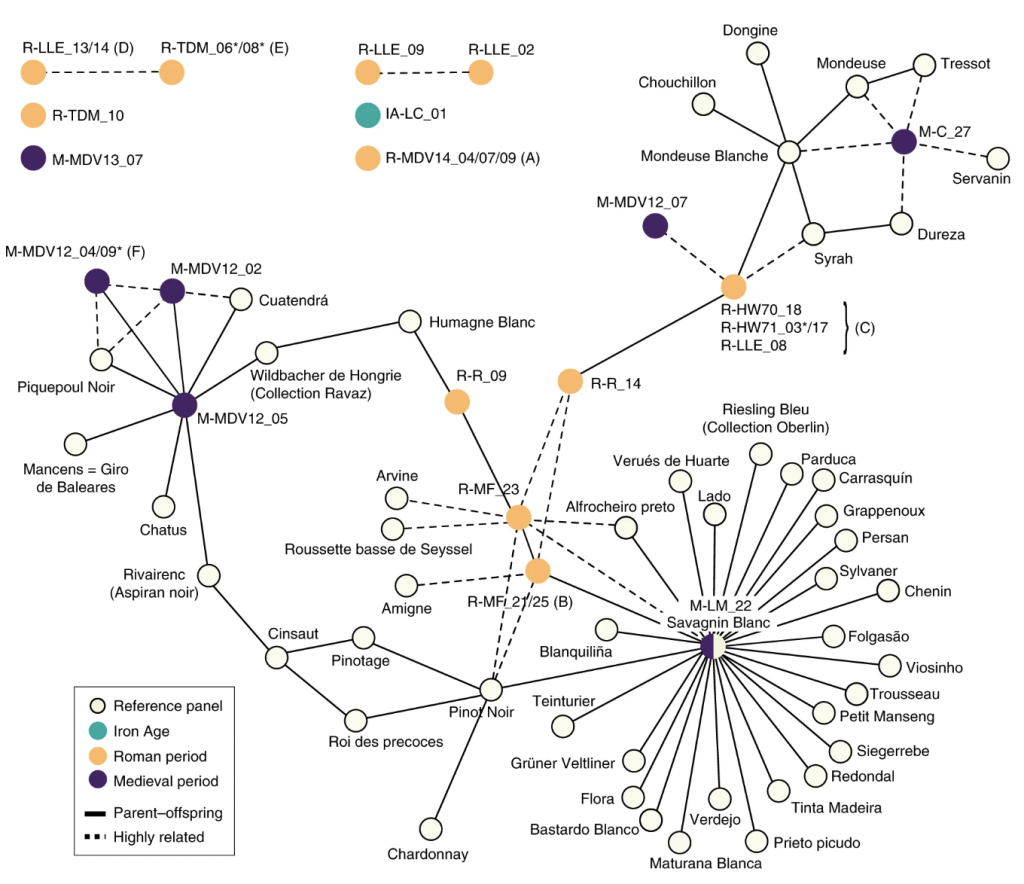 The grapevine has it that some living varieties of Vitis vinifera were grown by the Romans, and maintained for over two millennia. Ramos-Madrigal et al. used target-enriched genome-wide sequencing for 28 grape seeds recovered from archeological sites dating to the Iron Age, Roman period and Middle Ages. The meta-analysis revealed that these archeological grapes were more closely related to modern varieties from West and Central Europe than to wild grape relatives. By calculating the kinship coefficient between the ancient samples, the close relationships were found for archeological samples found in sites 600 km apart, suggesting that the grape cuttings were transported across long distances in ancient times. Additionally, some of the ancient varieties were also found to be similar to modern varieties, suggesting that varieties related to “Savagnin Blanc” were grown in France since approximately two millennia ago (Summarized by Magdalena Julkowska) Nature Plants 10.1038/s41477-019-0437-5
The grapevine has it that some living varieties of Vitis vinifera were grown by the Romans, and maintained for over two millennia. Ramos-Madrigal et al. used target-enriched genome-wide sequencing for 28 grape seeds recovered from archeological sites dating to the Iron Age, Roman period and Middle Ages. The meta-analysis revealed that these archeological grapes were more closely related to modern varieties from West and Central Europe than to wild grape relatives. By calculating the kinship coefficient between the ancient samples, the close relationships were found for archeological samples found in sites 600 km apart, suggesting that the grape cuttings were transported across long distances in ancient times. Additionally, some of the ancient varieties were also found to be similar to modern varieties, suggesting that varieties related to “Savagnin Blanc” were grown in France since approximately two millennia ago (Summarized by Magdalena Julkowska) Nature Plants 10.1038/s41477-019-0437-5
An Improved plant toolset for high-throughput recombineering (BioRxiv)
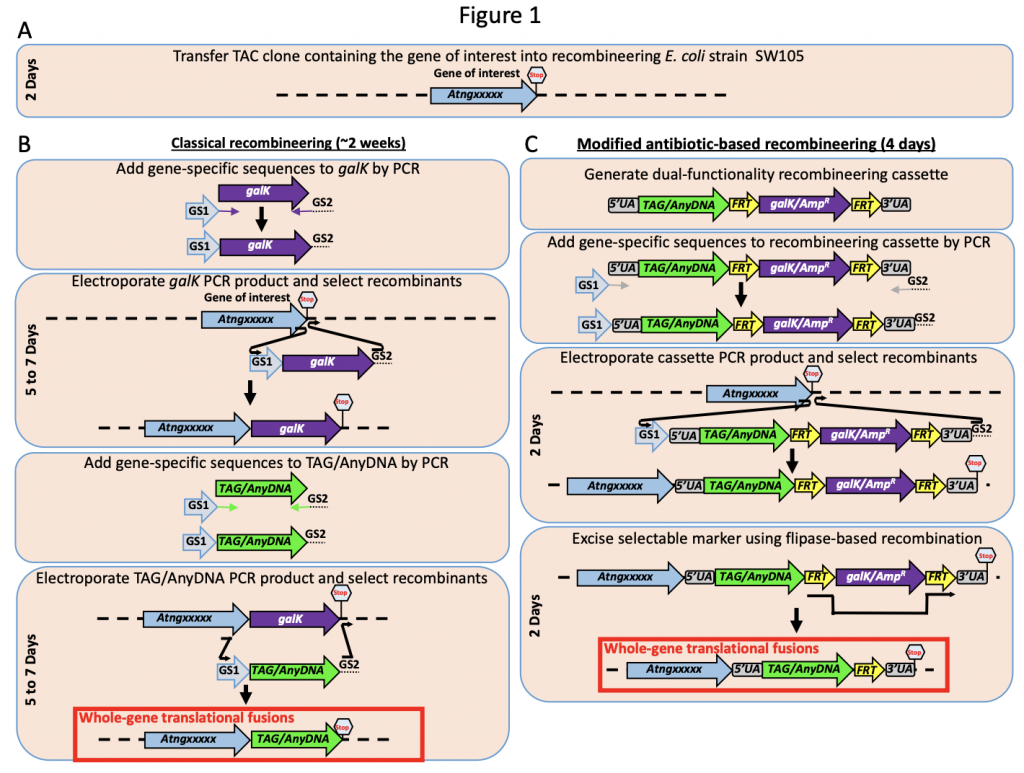 To determine the role of any gene of interest it is necessary to understand the spatiotemporal expression pattern. This is accomplished by tagging the regulatory sequence(s) alone (transcriptional fusion) or regulatory sequence(s) with the coding region (translational fusion) of the gene of interest. In this paper Brumos et al., have developed a recombineering based gene tagging approach that could efficiently tag the gene of interest. In this approach, the authors have created a recombinase-mediated cassette that is available in public stock centers including ABRC, which allows the transfer of sequence from a BAC (Bacterial Artificial Chromosome) clone in to plant transformation compatible binary vector. The efficiency of the system has been evaluated in Arabidopsis by tagging multiple genes involved in auxin-related genes but expandable beyond Arabidopsis. Thus the tool developed here provides more precise gene modifications and reduces the labor/time involved in generating these valuable constructs. (Summarized by Suresh Damodaran) bioRxiv 10.1101/659276
To determine the role of any gene of interest it is necessary to understand the spatiotemporal expression pattern. This is accomplished by tagging the regulatory sequence(s) alone (transcriptional fusion) or regulatory sequence(s) with the coding region (translational fusion) of the gene of interest. In this paper Brumos et al., have developed a recombineering based gene tagging approach that could efficiently tag the gene of interest. In this approach, the authors have created a recombinase-mediated cassette that is available in public stock centers including ABRC, which allows the transfer of sequence from a BAC (Bacterial Artificial Chromosome) clone in to plant transformation compatible binary vector. The efficiency of the system has been evaluated in Arabidopsis by tagging multiple genes involved in auxin-related genes but expandable beyond Arabidopsis. Thus the tool developed here provides more precise gene modifications and reduces the labor/time involved in generating these valuable constructs. (Summarized by Suresh Damodaran) bioRxiv 10.1101/659276
The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor (Nature Gen.)
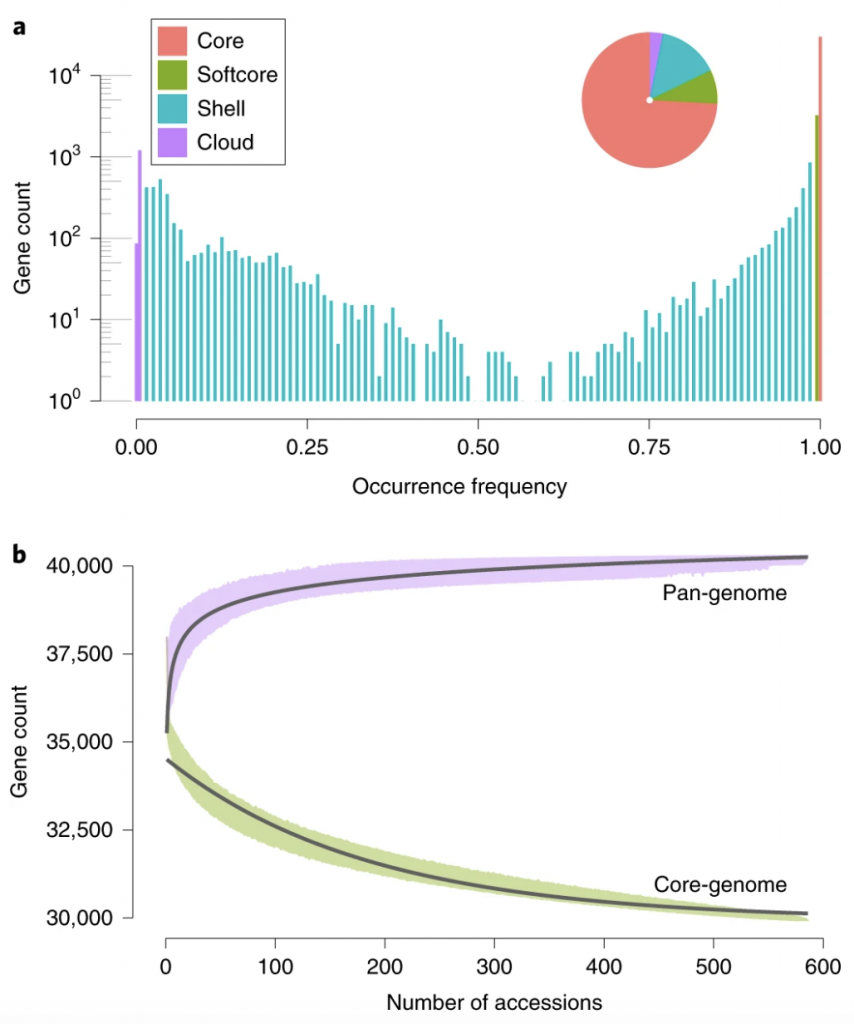 The wide variety of shape and flavor in modern cultivated tomatoes is a result of introgressions from its wild relatives. While many genetic studies were successful in the identification of wild alleles that affect tomato aroma, no tools were available to study structural variants affecting the taste, missing from the reference line. Gao et al. used the genomic sequences of large-fruit sized tomatoes, cherry tomatoes, and tomato’s closest wild relatives, and by de novo assembly was able to identify previously unknown sequences, not present in the reference genome. A total of 5,000 protein-coding genes were predicted in the sequences not represented in the reference genome. Additionally, almost 26% of genes in the pan-genome showed varying degree of presence/absence variation, indicating the diverse makeup of Lycopersidae clade. The wild tomato genomes were not only containing more genes, but they were enriched in defense responses, oxidation-reduction, and senescence. The pan-genome analysis enabled to identify a 4 kb substitution in the promoter region of TomLoxC, which was also mapped using a QTL study to be responsible for the variation in for volatiles and apocarotenoids production. (Summarized by Magdalena Julkowska) Nature Genetics 10.1038/s41588-019-0410-2
The wide variety of shape and flavor in modern cultivated tomatoes is a result of introgressions from its wild relatives. While many genetic studies were successful in the identification of wild alleles that affect tomato aroma, no tools were available to study structural variants affecting the taste, missing from the reference line. Gao et al. used the genomic sequences of large-fruit sized tomatoes, cherry tomatoes, and tomato’s closest wild relatives, and by de novo assembly was able to identify previously unknown sequences, not present in the reference genome. A total of 5,000 protein-coding genes were predicted in the sequences not represented in the reference genome. Additionally, almost 26% of genes in the pan-genome showed varying degree of presence/absence variation, indicating the diverse makeup of Lycopersidae clade. The wild tomato genomes were not only containing more genes, but they were enriched in defense responses, oxidation-reduction, and senescence. The pan-genome analysis enabled to identify a 4 kb substitution in the promoter region of TomLoxC, which was also mapped using a QTL study to be responsible for the variation in for volatiles and apocarotenoids production. (Summarized by Magdalena Julkowska) Nature Genetics 10.1038/s41588-019-0410-2