Plant Science Research Weekly: June 19th
Review Single-cell genomics and epigenomics: Technologies and applications in plants ($)
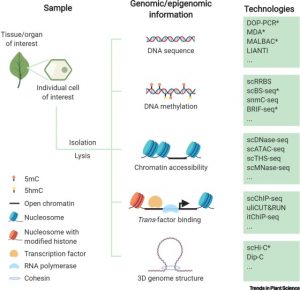 Plants (embryophytes) are by definition multicellular, but we seek to understand them as the sum of the activities of individual cells. Much of this knowledge rests on information obtained through grinding up tissues made up from several cell types. This review by Luo et al. describes methods for plant single-cell genomics and epigenomics studies. All involve first isolating single cells, followed by whole-genome amplification to obtain sufficient material for analysis. As many of the techniques were developed for use with easy-to-disperse animal cells, the additional steps required to dissociate plant cells adds potential for artifacts. At the genomic sequence level, single-cell methods can point to single-nucleotide as well as copy-number variations. A suite of epigenomic analysis are also possible, including analysis of DNA methylation and even DNA-histone interactions, though as yet it isn’t possible to survey transcription factor binding within single cells. The authors describe some applications of these technologies, and also point to the promise of complete -omics datasets for every cell type. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.04.016
Plants (embryophytes) are by definition multicellular, but we seek to understand them as the sum of the activities of individual cells. Much of this knowledge rests on information obtained through grinding up tissues made up from several cell types. This review by Luo et al. describes methods for plant single-cell genomics and epigenomics studies. All involve first isolating single cells, followed by whole-genome amplification to obtain sufficient material for analysis. As many of the techniques were developed for use with easy-to-disperse animal cells, the additional steps required to dissociate plant cells adds potential for artifacts. At the genomic sequence level, single-cell methods can point to single-nucleotide as well as copy-number variations. A suite of epigenomic analysis are also possible, including analysis of DNA methylation and even DNA-histone interactions, though as yet it isn’t possible to survey transcription factor binding within single cells. The authors describe some applications of these technologies, and also point to the promise of complete -omics datasets for every cell type. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.04.016
RALF1-FERONIA complex affects splicing dynamics to modulate stress responses and growth in plants
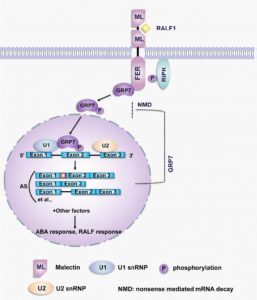 Alternative splicing is a process that can produce two or more transcript isoforms from a single gene and can increase protein diversity. FERONIA (FER) is a receptor-like kinase that serves as a receptor for the rapid alkalinization factor (RALF) peptides that regulate multiple cellular activities in response to biotic and abiotic stresses. Wang et al. investigated signalling downstream of RALF and FER. Upon RALF1 treatment, more than 3000 genes show alternative splicing events, including many that are stress-regulated or involved in amino acid biosynthesis. The authors screened for proteins that interact with the FER receptor and identified a splicing factor glycine-rich RNA binding protein7 (GRP7). FER interacts with and phosphorylates GPR7 in a RALF-dependent manner. Phosphorylated GRP7 translocates into the nucleus and shows increased RNA-binding activity to its target mRNAs. Upon ABA treatment, fer-4 and grp7 mutants show stronger root inhibition than the wild type, indicating that they play a negative role in ABA responses. Interestingly, ABA treatment leads to an increase of alternatively-spliced GRP7 transcripts, and induces its degradation via nonsense-mediated mRNA decay (NMD), suggesting a mechanism for fine-tuning this pathway. (Summary by Min May Wong @WongMinMay) Science Advances 10.1126/sciadv.aaz1622
Alternative splicing is a process that can produce two or more transcript isoforms from a single gene and can increase protein diversity. FERONIA (FER) is a receptor-like kinase that serves as a receptor for the rapid alkalinization factor (RALF) peptides that regulate multiple cellular activities in response to biotic and abiotic stresses. Wang et al. investigated signalling downstream of RALF and FER. Upon RALF1 treatment, more than 3000 genes show alternative splicing events, including many that are stress-regulated or involved in amino acid biosynthesis. The authors screened for proteins that interact with the FER receptor and identified a splicing factor glycine-rich RNA binding protein7 (GRP7). FER interacts with and phosphorylates GPR7 in a RALF-dependent manner. Phosphorylated GRP7 translocates into the nucleus and shows increased RNA-binding activity to its target mRNAs. Upon ABA treatment, fer-4 and grp7 mutants show stronger root inhibition than the wild type, indicating that they play a negative role in ABA responses. Interestingly, ABA treatment leads to an increase of alternatively-spliced GRP7 transcripts, and induces its degradation via nonsense-mediated mRNA decay (NMD), suggesting a mechanism for fine-tuning this pathway. (Summary by Min May Wong @WongMinMay) Science Advances 10.1126/sciadv.aaz1622
Divide to heal: auxin regulates wound-induced cell division in roots
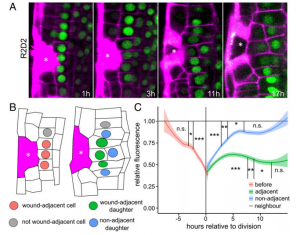 Among the stresses that sessile plants face, wounding is peculiar as it can be caused by both biotic and abiotic sources. While studies of wounding in animal systems have largely focused on the repair mechanisms, less is known about wound healing in plants. In an attempt to throw light on this, Hoermayer and colleagues have characterized auxin as an important regulator of wound healing in root tissues. The authors used UV laser ablation for targeted wounding of root cells, which induced a steep rise in auxin concentration in the cells immediately adjacent to the ablated cells. Using pharmacological agents, they showed this auxin signaling is required for cell expansion and tissue regeneration after wounding. Genetic studies revealed that the perception of auxin is routed through the canonical auxin receptors TIR1/AFBs and the signal is transduced through the wound-inducible ERF115 transcription factor, which accumulates in the adjacent cells. Interestingly, genetic disruption of TIR1 leads to tumor-like over-proliferation of tissue around the wound site, demonstrating the importance of canonical auxin response pathway in regulating the responses. Apart from genetic factors, the authors also found physiological elements like turgor pressure, generated due to disruption of wounded cells, play a role in inducing the subsequent processes of regeneration. (Summary by Pavithran Narayanan @pavi_narayanan) Proc. Natl. Acad. Sci. USA
Among the stresses that sessile plants face, wounding is peculiar as it can be caused by both biotic and abiotic sources. While studies of wounding in animal systems have largely focused on the repair mechanisms, less is known about wound healing in plants. In an attempt to throw light on this, Hoermayer and colleagues have characterized auxin as an important regulator of wound healing in root tissues. The authors used UV laser ablation for targeted wounding of root cells, which induced a steep rise in auxin concentration in the cells immediately adjacent to the ablated cells. Using pharmacological agents, they showed this auxin signaling is required for cell expansion and tissue regeneration after wounding. Genetic studies revealed that the perception of auxin is routed through the canonical auxin receptors TIR1/AFBs and the signal is transduced through the wound-inducible ERF115 transcription factor, which accumulates in the adjacent cells. Interestingly, genetic disruption of TIR1 leads to tumor-like over-proliferation of tissue around the wound site, demonstrating the importance of canonical auxin response pathway in regulating the responses. Apart from genetic factors, the authors also found physiological elements like turgor pressure, generated due to disruption of wounded cells, play a role in inducing the subsequent processes of regeneration. (Summary by Pavithran Narayanan @pavi_narayanan) Proc. Natl. Acad. Sci. USA
Dual-sensing genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic ABA and second messenger dynamics
 Responses to a changing environment require an interplay between primary and secondary messengers. Hormones like abscisic acid (ABA), auxin etc. act as a primary messengers, whereas molecules like Ca2+, reactive oxygen species (ROS), other cations and anions act as a secondary messenger. In this paper, Waadt et al. generated a set of dual-reporting genetically encoded fluorescent indicators (2-in-1 GEFIs) that allow observation of the dynamics of two messengers at the same time in Arabidopsis roots. The indicators and ligands they recognize (in parenthesis) are: ABAleonSD1-3L21 (ABA), R-GECO1 (Ca2+), Arabidopsis codon optimized red-fluorescing (P)A-17 (H+), E2GFP (H+ and Cl–), Grx1-roGFP2 (glutathione redox potential; EGSH) and roGFP2-Orp1 (H2O2). These were linked by either a 14 amino acid peptide or a self-cleaving P2A linker that allows the two indicators to be expressed separately but in equal amounts. The sensors were imaged in different combinations, which showed the dynamics of ABA, Ca2+, H+ and H2O2 in response to various chemicals including ABA, IAA, glutamate, ATP, and H2O2. Thus, this paper reveals the interdependence of each signaling molecule and provides new tools for high resolution spatiotemporal information. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant Cell 10.1105/tpc.19.00892
Responses to a changing environment require an interplay between primary and secondary messengers. Hormones like abscisic acid (ABA), auxin etc. act as a primary messengers, whereas molecules like Ca2+, reactive oxygen species (ROS), other cations and anions act as a secondary messenger. In this paper, Waadt et al. generated a set of dual-reporting genetically encoded fluorescent indicators (2-in-1 GEFIs) that allow observation of the dynamics of two messengers at the same time in Arabidopsis roots. The indicators and ligands they recognize (in parenthesis) are: ABAleonSD1-3L21 (ABA), R-GECO1 (Ca2+), Arabidopsis codon optimized red-fluorescing (P)A-17 (H+), E2GFP (H+ and Cl–), Grx1-roGFP2 (glutathione redox potential; EGSH) and roGFP2-Orp1 (H2O2). These were linked by either a 14 amino acid peptide or a self-cleaving P2A linker that allows the two indicators to be expressed separately but in equal amounts. The sensors were imaged in different combinations, which showed the dynamics of ABA, Ca2+, H+ and H2O2 in response to various chemicals including ABA, IAA, glutamate, ATP, and H2O2. Thus, this paper reveals the interdependence of each signaling molecule and provides new tools for high resolution spatiotemporal information. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant Cell 10.1105/tpc.19.00892
A FRET sensor reports kinase activity in planta
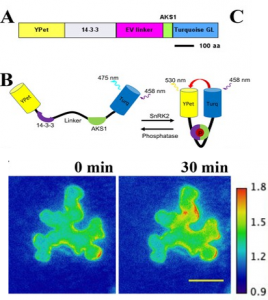 Abscisic acid (ABA) is a phytohormone involved in imparting tolerance to a variety of stress conditions. Importantly, ABA is involved in stomatal closure, mediated by the sucrose non-fermenting1-related protein kinase 2 (SnRK2) family of proteins. To report SnRK2 activity in vivo, Zhang and co-workers developed a FRET-based fluorescent sensor (named SNACs for SnRK2 activity sensor). This sensor exploits a domain from a natural substrate of SnRK2s and a 14-3-3 domain protein that responds to phosphorylation of the substrate protein. When SnRK2 phosphorylates the substrate protein, conformation of the sensor changes to induce a Förster resonance energy transfer (FRET) between the fluorophores, resulting in fluorescence emission. The sensor reported SnRK2 activation by ABA, but high ambient CO2 and external MeJA application failed to induce SnRK2 kinase activity, suggesting that stomatal CO2 signaling requires basal ABA and SnRK2 signaling, but not SnRK2 activation. This report is expected to fuel development of sensors for other phosphorylation-based processes and studies to determine the pathway(s) involved in CO2 and MeJA-induced stomatal closure. (Summary by Pavithran Narayanan @pavi_narayanan) eLIFE 10.7554/eLife.56351
Abscisic acid (ABA) is a phytohormone involved in imparting tolerance to a variety of stress conditions. Importantly, ABA is involved in stomatal closure, mediated by the sucrose non-fermenting1-related protein kinase 2 (SnRK2) family of proteins. To report SnRK2 activity in vivo, Zhang and co-workers developed a FRET-based fluorescent sensor (named SNACs for SnRK2 activity sensor). This sensor exploits a domain from a natural substrate of SnRK2s and a 14-3-3 domain protein that responds to phosphorylation of the substrate protein. When SnRK2 phosphorylates the substrate protein, conformation of the sensor changes to induce a Förster resonance energy transfer (FRET) between the fluorophores, resulting in fluorescence emission. The sensor reported SnRK2 activation by ABA, but high ambient CO2 and external MeJA application failed to induce SnRK2 kinase activity, suggesting that stomatal CO2 signaling requires basal ABA and SnRK2 signaling, but not SnRK2 activation. This report is expected to fuel development of sensors for other phosphorylation-based processes and studies to determine the pathway(s) involved in CO2 and MeJA-induced stomatal closure. (Summary by Pavithran Narayanan @pavi_narayanan) eLIFE 10.7554/eLife.56351
Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars
 Crop production is greatly influenced by the duration of the last stage of plant life cycle, senescence, through degradation of resources in leaves and remobilization of nutrients to developing seeds. Indeed, higher grain yield of important cereals such as maize and sorghum can be achieved by using stay-green (sgr) mutants characterized by delayed senescence. Nevertheless, this agronomical trait has not been fully explored in Asian rice (O. sativa). In this article, Shin and colleagues investigated the genetic basis of senescence by crossing cultivars belonging to the subspecies japonica and indica, which show similar heading dates but different lifespans. QTL mapping revealed that differential senescence correlated with variations in the promoter region of the OsSGR gene, encoding an enzyme involved in chlorophyll catabolism. Polymorphisms in the indica allele associated with earlier activation and higher expression of OsSGR, leading to a faster chlorophyll breakdown, reduced photosynthetic ability and decreased yield. Further analyses suggested that senescence patterns of rice varieties have been selected on the basis of the growing areas and cultivation practices: indica cultivars are better suited for tropical regions with double or triple cropping systems, whereas japonica cultivars are better suited for temperate and subtropical regions with a single harvesting system. Last, the authors generated Near Isogenic Lines (NIL) carrying the japonica OsSGR alleles in indica genetic background, and observed delayed senescence and an average 10% increase in grain yield. These findings indicate that prolonged photosynthetic ability and higher chlorophyll content can improve grain filling and crop production. (Summary by Michela Osnato @michela_osnato) Nature Comms. 10.1038/s41467-020-16573-2
Crop production is greatly influenced by the duration of the last stage of plant life cycle, senescence, through degradation of resources in leaves and remobilization of nutrients to developing seeds. Indeed, higher grain yield of important cereals such as maize and sorghum can be achieved by using stay-green (sgr) mutants characterized by delayed senescence. Nevertheless, this agronomical trait has not been fully explored in Asian rice (O. sativa). In this article, Shin and colleagues investigated the genetic basis of senescence by crossing cultivars belonging to the subspecies japonica and indica, which show similar heading dates but different lifespans. QTL mapping revealed that differential senescence correlated with variations in the promoter region of the OsSGR gene, encoding an enzyme involved in chlorophyll catabolism. Polymorphisms in the indica allele associated with earlier activation and higher expression of OsSGR, leading to a faster chlorophyll breakdown, reduced photosynthetic ability and decreased yield. Further analyses suggested that senescence patterns of rice varieties have been selected on the basis of the growing areas and cultivation practices: indica cultivars are better suited for tropical regions with double or triple cropping systems, whereas japonica cultivars are better suited for temperate and subtropical regions with a single harvesting system. Last, the authors generated Near Isogenic Lines (NIL) carrying the japonica OsSGR alleles in indica genetic background, and observed delayed senescence and an average 10% increase in grain yield. These findings indicate that prolonged photosynthetic ability and higher chlorophyll content can improve grain filling and crop production. (Summary by Michela Osnato @michela_osnato) Nature Comms. 10.1038/s41467-020-16573-2
Robotic Assay for Drought (RoAD): An automated phenotyping system for brassinosteroid and drought responses
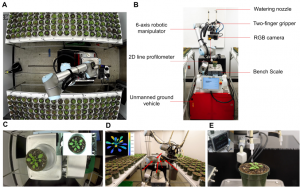
Developing drought tolerant plants is an important challenge in agriculture. Drought responses and plant growth are regulated by several signaling pathways, one of which is activated by brassinosteroids (BRs), a group of plant steroid hormones. In order to better understand the crosstalk between BR-mediated growth and drought responses, Xiang et al. developed the Robotic Assay for Drought (RoAD), a robotic phenotyping platform that obtains 2D and 3D images to measure morphological traits. The system is capable of watering and maintaining plants at different soil moisture conditions to quantify growth responses of different lines and treatments under different drought conditions. The system also incorporates an automatic image processing pipeline, supporting plant and leaf segmentation, and calculation of morphological and color features. Plants of Arabidopsis and maize treated with propiconazole (a BR biosynthesis inhibitor) showed more compact growth compared to the control, as did bril-301, a loss-of-function Arabidopsis BR mutant receptor. Furthermore, different Arabidopsis accessions showed different responses to propiconazole, providing new opportunities to explore the coordination between BR-mediated growth and stress responses. (Summary by Elisandra Pradella @Elisandra_MP) bioRxiv 10.1101/2020.06.01.128199
Co‐catabolism of arginine and succinate drives symbiotic nitrogen fixation
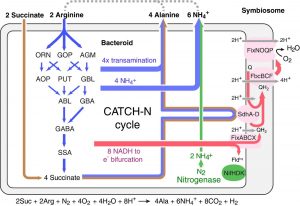 Symbiotic nitrogen fixation is a mutualistic relationship between plants and microbes in which plants supply fixed carbon to bacteria in exchange for nitrogen. During this process, the microbes use nitrogenase enzyme to convert atmospheric nitrogen into a plant-usable form, but the metabolic interaction between plants and microbes in maintaining this energy-intensive nitrogenase reaction remains an open question. In this paper, Tinoco et al. found that co-supply of both succinate (a carbon source) and arginine (a nitrogen source) are required to sustain the energy-intensive nitrogen fixation process. Co-feeding of succinate and arginine to isolated mature bacteroids of Bradyrhizobium diazoefficiens and Sinorrhizium meliloti increased the ATP levels and nitrogenase enzyme activity to a similar extent as providing the bacteroids with nodule crude extracts. To identify the pathway, the authors used isotope labeling and in planta transposon sequencing, in which a transposon-mutagenized pool of bacteria was introduced into plants and recovered after six weeks of in planta selection; genes underrepresented in the recovered sample are involved in symbiosis. These studies showed that arginine transport and catabolism are required during symbiosis, which was further validated using S. meliloti mutants impaired in arginine transport and catabolism. The authors named this newly identified metabolic pathway CATCH-N (C4‐dicarboxylate arginine transamination co‐catabolism under acidic (H+) conditions to fix nitrogen), and showed that the CATCH-N cycle delivers a 25% increase in nitrogen as well as promotes the survival of bacteria within the symbiosome. This cycle provides an important opportunity for engineering improved nitrogen-fixing symbiosis. (Summary by Sunita Pratak @psunita) Mol. Sys. Biol. 10.15252/msb.20199419
Symbiotic nitrogen fixation is a mutualistic relationship between plants and microbes in which plants supply fixed carbon to bacteria in exchange for nitrogen. During this process, the microbes use nitrogenase enzyme to convert atmospheric nitrogen into a plant-usable form, but the metabolic interaction between plants and microbes in maintaining this energy-intensive nitrogenase reaction remains an open question. In this paper, Tinoco et al. found that co-supply of both succinate (a carbon source) and arginine (a nitrogen source) are required to sustain the energy-intensive nitrogen fixation process. Co-feeding of succinate and arginine to isolated mature bacteroids of Bradyrhizobium diazoefficiens and Sinorrhizium meliloti increased the ATP levels and nitrogenase enzyme activity to a similar extent as providing the bacteroids with nodule crude extracts. To identify the pathway, the authors used isotope labeling and in planta transposon sequencing, in which a transposon-mutagenized pool of bacteria was introduced into plants and recovered after six weeks of in planta selection; genes underrepresented in the recovered sample are involved in symbiosis. These studies showed that arginine transport and catabolism are required during symbiosis, which was further validated using S. meliloti mutants impaired in arginine transport and catabolism. The authors named this newly identified metabolic pathway CATCH-N (C4‐dicarboxylate arginine transamination co‐catabolism under acidic (H+) conditions to fix nitrogen), and showed that the CATCH-N cycle delivers a 25% increase in nitrogen as well as promotes the survival of bacteria within the symbiosome. This cycle provides an important opportunity for engineering improved nitrogen-fixing symbiosis. (Summary by Sunita Pratak @psunita) Mol. Sys. Biol. 10.15252/msb.20199419
Influential neighbours: Seeds of dominant species affect the germination of common grassland species
 Seed-seed interactions can control the germination of grassland species. The result of this interaction (i.e., germination being inhibited or promoted) presumably depends on the dominance and taxonomic relatedness of interacting species. However, the relative importance of these factors is poorly explored. Here, Fenesi et al. assessed the interactions between seeds of nine perennial species from a semi-dry grassland, focusing on the effect of dominant and subordinate species on the germination of three Asteraceae target plants. The authors incubated seeds of the target species with varying densities of a neighboring species seeds. They also tested the effect of water availability on these interactions. Regardless of taxonomic relatedness, growing along dominant species’ seeds had a mixed impact on the germination of target species, being positive, negative, or null depending on the target species. Also, the density of neighboring seeds and water availability changed magnitude the direction of the effect, being positive in most high-density treatments and negative in low-density ones. Given these results, the authors highlight the complexity of seed-seed interactions. Still, they support that the relative dominance of a species in an ecosystem has a more substantial influence on these than taxonomic relatedness of interacting species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Veg. Sci. 10.1111/jvs.12892
Seed-seed interactions can control the germination of grassland species. The result of this interaction (i.e., germination being inhibited or promoted) presumably depends on the dominance and taxonomic relatedness of interacting species. However, the relative importance of these factors is poorly explored. Here, Fenesi et al. assessed the interactions between seeds of nine perennial species from a semi-dry grassland, focusing on the effect of dominant and subordinate species on the germination of three Asteraceae target plants. The authors incubated seeds of the target species with varying densities of a neighboring species seeds. They also tested the effect of water availability on these interactions. Regardless of taxonomic relatedness, growing along dominant species’ seeds had a mixed impact on the germination of target species, being positive, negative, or null depending on the target species. Also, the density of neighboring seeds and water availability changed magnitude the direction of the effect, being positive in most high-density treatments and negative in low-density ones. Given these results, the authors highlight the complexity of seed-seed interactions. Still, they support that the relative dominance of a species in an ecosystem has a more substantial influence on these than taxonomic relatedness of interacting species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Veg. Sci. 10.1111/jvs.12892
An aphid RNA transcript migrates systemically within plants and is a virulence factor
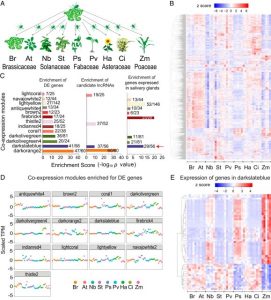 Aphids are an important insect pest that can cause significant yield loss to crops, particularly through feeding damage and as vector for devasting plant pathogens. How aphids and other sap-feeding insects use their stylet to penetrate the plant vascular tissues is well studied but the factors involved in establishing a long-term interaction between aphids and plant and how this occurs remain largely unknown. Chen et al. demonstrated that the green peach aphid M. persicae can colonize nine plant species from different families indicating that it is a true generalist. RNA-seq analysis using samples from nine host plants showed that M. persicae can coordinate its gene expression in response to the plant species it colonizes. Further analysis showed that differentially expressed genes are enriched for candidate long noncoding RNA (lncRNA) and some of the genes lie adjacent to each other on the aphid genome as tandem repeats. Analysis also revealed that some of these lncRNA belong to the same gene family and can translocate to distal leaves; the authors named them the Ya family (Ya means aphid in Chinese). Stable expression of M. persicae Ya1 RNA increases aphid reproduction whereas their fecundity decreases in RNAi lines, suggesting that YA1 is a virulence factor. This study showed that beyond serving as a vector for plant pathogens, M. persicae translocates it own RNA transcripts into the host plant as virulence factors. (Summary by Toluwase Olukayodo @toluxylic) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1918410117
Aphids are an important insect pest that can cause significant yield loss to crops, particularly through feeding damage and as vector for devasting plant pathogens. How aphids and other sap-feeding insects use their stylet to penetrate the plant vascular tissues is well studied but the factors involved in establishing a long-term interaction between aphids and plant and how this occurs remain largely unknown. Chen et al. demonstrated that the green peach aphid M. persicae can colonize nine plant species from different families indicating that it is a true generalist. RNA-seq analysis using samples from nine host plants showed that M. persicae can coordinate its gene expression in response to the plant species it colonizes. Further analysis showed that differentially expressed genes are enriched for candidate long noncoding RNA (lncRNA) and some of the genes lie adjacent to each other on the aphid genome as tandem repeats. Analysis also revealed that some of these lncRNA belong to the same gene family and can translocate to distal leaves; the authors named them the Ya family (Ya means aphid in Chinese). Stable expression of M. persicae Ya1 RNA increases aphid reproduction whereas their fecundity decreases in RNAi lines, suggesting that YA1 is a virulence factor. This study showed that beyond serving as a vector for plant pathogens, M. persicae translocates it own RNA transcripts into the host plant as virulence factors. (Summary by Toluwase Olukayodo @toluxylic) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1918410117
Flowering plant composition shapes pathogen infection intensity and reproduction in bumble bee colonies
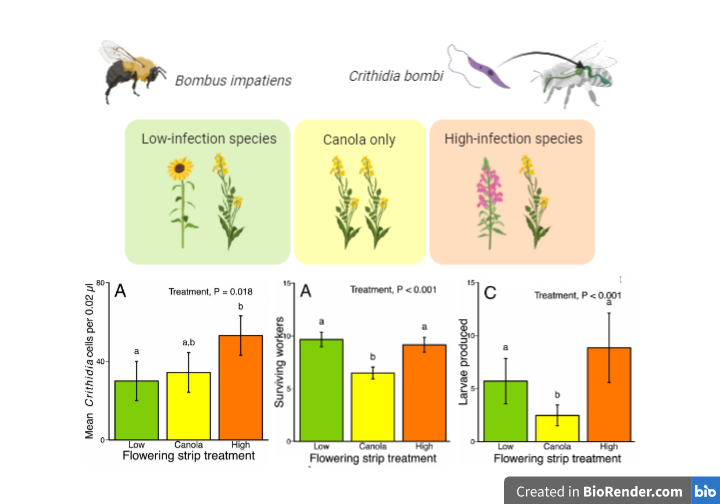 Pathogens are one of many factors underlying pollinator decline. Diseases can be transmitted from commercial honeybees to wild bees through flowers. Bumble bees (Bombus impatiens) are infected by the gut pathogen Crithidia bombi which is contracted at flowers by fecal-oral transmission. However, not all species of plants result in equal transmission of disease to bees, therefore Adler et al. sought to determine how species composition would affect pathogen infection and bumble bee colony fitness. They created tents of canola interspersed with flowering strips containing low- or high-infection plant species, or no strips, and spray inoculated C. bombi onto the plants before introducing the bees. Flowering strips containing high-infection species had double the infection intensity compared to low-infection. However, nectar availability, larva production, and worker survival were higher in both high- and low-infection plots compared to canola only, demonstrating the importance of flowering strips for bee health. This work can help inform planting pollinator friendly habitats to promote native been population while selecting plant species that minimize pathogen transmission. (Summary and image by Katy Dunning @PlantMomKaty) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2000074117
Pathogens are one of many factors underlying pollinator decline. Diseases can be transmitted from commercial honeybees to wild bees through flowers. Bumble bees (Bombus impatiens) are infected by the gut pathogen Crithidia bombi which is contracted at flowers by fecal-oral transmission. However, not all species of plants result in equal transmission of disease to bees, therefore Adler et al. sought to determine how species composition would affect pathogen infection and bumble bee colony fitness. They created tents of canola interspersed with flowering strips containing low- or high-infection plant species, or no strips, and spray inoculated C. bombi onto the plants before introducing the bees. Flowering strips containing high-infection species had double the infection intensity compared to low-infection. However, nectar availability, larva production, and worker survival were higher in both high- and low-infection plots compared to canola only, demonstrating the importance of flowering strips for bee health. This work can help inform planting pollinator friendly habitats to promote native been population while selecting plant species that minimize pathogen transmission. (Summary and image by Katy Dunning @PlantMomKaty) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2000074117
Plant genes for defense against insect eggs
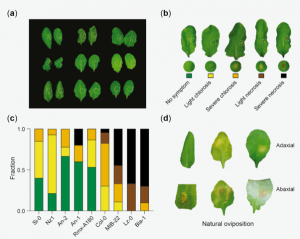 Plants can recognize eggs of herbivorous insects deposited on leaves and mount defense responses to avoid the future threat of herbivory. Similar to plant hypersensitive response against pathogens, plants can cause localized cell death to defend themselves against insect eggs. This response is called a hypersensitive-like response (HR-like). Despite similarities between HR-like and well-studied plant immune responses against pathogens, the molecular mechanisms underlying HR-like is poorly understood. Groux et al. exploited Arabidopsis natural variation in HR-like symptoms induced by egg extracts (EE) of the large white butterfly Pieris brassicae to carry out genome-wide association study (GWAS). The screening identified a strong association between HR-like symptoms and a locus containing L-TYPE LECTIN RECEPTOR KINASE-I.1 (LecRK-I.1), a previously uncharacterized gene for plant defense. Reverse genetics analysis confirmed that LecRK-I.1 is involved in EE-induced cell death. The authors found two major haplotypes of LecRK-I.1 broadly distributed across accessions and showed that this locus is under balancing selection, a type of selection in which genetic variation is maintained in a population and is commonly observed in immunity-related genes such as immune receptors. The result suggests that LecRK-I.1 might contribute to plant adaptation to specific environments. This study deepens our understanding of parallels between plant defense systems against insect eggs and pathogens and sets the stage for mechanistic interrogation of the enigmatic plant HR-like response. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.29.122846
Plants can recognize eggs of herbivorous insects deposited on leaves and mount defense responses to avoid the future threat of herbivory. Similar to plant hypersensitive response against pathogens, plants can cause localized cell death to defend themselves against insect eggs. This response is called a hypersensitive-like response (HR-like). Despite similarities between HR-like and well-studied plant immune responses against pathogens, the molecular mechanisms underlying HR-like is poorly understood. Groux et al. exploited Arabidopsis natural variation in HR-like symptoms induced by egg extracts (EE) of the large white butterfly Pieris brassicae to carry out genome-wide association study (GWAS). The screening identified a strong association between HR-like symptoms and a locus containing L-TYPE LECTIN RECEPTOR KINASE-I.1 (LecRK-I.1), a previously uncharacterized gene for plant defense. Reverse genetics analysis confirmed that LecRK-I.1 is involved in EE-induced cell death. The authors found two major haplotypes of LecRK-I.1 broadly distributed across accessions and showed that this locus is under balancing selection, a type of selection in which genetic variation is maintained in a population and is commonly observed in immunity-related genes such as immune receptors. The result suggests that LecRK-I.1 might contribute to plant adaptation to specific environments. This study deepens our understanding of parallels between plant defense systems against insect eggs and pathogens and sets the stage for mechanistic interrogation of the enigmatic plant HR-like response. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2020.05.29.122846