Plant Science Research Weekly: June 17, 2022
Review: Molecular regulators of nitrate response in plants
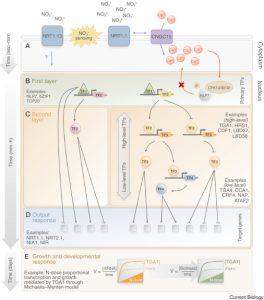 Nitrate is the major form of nitrogen used by plants in an aerobic crop cultivation scenario. Lamig et al. review recent additions to the already vast knowledge of nitrate signaling. A first line of regulation concerns nitrate uptake through post-transcriptional and post-translational regulation of nitrate transporters. Both low affinity (NRT1) and high affinity (NRT2) nitrate transporters are regulated by specific phosphorylation events determining their affinity or functionality. For example, Ser 501 phosphorylation suppresses NRT2.1 mediated nitrate transport under high nitrate supply. Many exciting new developments concern how information about nitrate availability regulates downstream events. Nitrate transporters are known as “transceptors” as they act as both transporters of nitrate and receptors that trigger downstream processes. (Furthermore, NRT1.1 can also transport auxin.) A recent study reports that under low nitrate conditions the cyclic nucleotide gated channel CNGC15 forms a heterocomplex with NRT1.1 and is inactivated, whereas under high nitrate conditions CNGC15 disassociates from NRT1.1 and functions as a calcium channel. Other processes are under the control of nitrate-responsive transcription factors. For example, the nitrate dependent nuclear retention of NLP7 regulates the expression of around 60% of nitrate induced genes. However, NLP7 is just one transcription factor within a large and complex regulatory hierarchy. The review also explores the mechanisms through which nitrate fine-tunes early plant development, including cotyledon and leaf growth and root and shoot architecture. Further studies are needed to address organ specific responses to nitrate, long term nitrate signaling, and interaction of nitrate signaling with other environmental factors. This expansive understanding of nitrate signaling and plant growth, mediated by the cooperation of multiple transcription factors, will aid in improving crop nitrogen use efficiency (Summary by Lekshmy Sathee @lekshmysnair) Curr. Biol. 10.1016/j.cub.2022.03.022
Nitrate is the major form of nitrogen used by plants in an aerobic crop cultivation scenario. Lamig et al. review recent additions to the already vast knowledge of nitrate signaling. A first line of regulation concerns nitrate uptake through post-transcriptional and post-translational regulation of nitrate transporters. Both low affinity (NRT1) and high affinity (NRT2) nitrate transporters are regulated by specific phosphorylation events determining their affinity or functionality. For example, Ser 501 phosphorylation suppresses NRT2.1 mediated nitrate transport under high nitrate supply. Many exciting new developments concern how information about nitrate availability regulates downstream events. Nitrate transporters are known as “transceptors” as they act as both transporters of nitrate and receptors that trigger downstream processes. (Furthermore, NRT1.1 can also transport auxin.) A recent study reports that under low nitrate conditions the cyclic nucleotide gated channel CNGC15 forms a heterocomplex with NRT1.1 and is inactivated, whereas under high nitrate conditions CNGC15 disassociates from NRT1.1 and functions as a calcium channel. Other processes are under the control of nitrate-responsive transcription factors. For example, the nitrate dependent nuclear retention of NLP7 regulates the expression of around 60% of nitrate induced genes. However, NLP7 is just one transcription factor within a large and complex regulatory hierarchy. The review also explores the mechanisms through which nitrate fine-tunes early plant development, including cotyledon and leaf growth and root and shoot architecture. Further studies are needed to address organ specific responses to nitrate, long term nitrate signaling, and interaction of nitrate signaling with other environmental factors. This expansive understanding of nitrate signaling and plant growth, mediated by the cooperation of multiple transcription factors, will aid in improving crop nitrogen use efficiency (Summary by Lekshmy Sathee @lekshmysnair) Curr. Biol. 10.1016/j.cub.2022.03.022
Upstream ORFs check undesired leaky protein translation and buffer noise
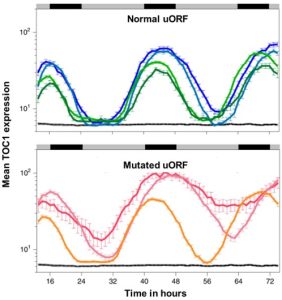 During my research journey, I have come across a handful of genes that are hard to express. Sometimes, even with a strong promoter and high transcript level, the protein level is negligible. We generally considered these proteins might be degrading very fast. But maybe not. A recent article by Wu et al. indicates translation of genes could be controlled by an upstream open reading frame (uORF). uORFs are upstream elements in the transcript that regulate the translation of about 30-50% of genes. The authors found that genes lacking uORF show leaky protein expression but the genes that have uORF don’t show this leaky expression. The authors observed the presence of uORF in a circadian-regulated gene TOC1. They mutated the uORF of TOC1 and observed that the mutated version produced more protein per mRNA than TOC1 harboring the normal uORF, indicating this uORF reduces protein expression. The authors also observed that the lack of proper uORF in Arabidopsis cells leads to loss of rhythmicity of TOC1 protein level, and that the uORF decreases cell-to-cell variability in TOC1 rhythmicity; that by buffering the expression level, noise is also reduced. They created a delightful animation that explains how this works. Overall, uORF elements seem to check undesired protein expression. Looking at the transcript of my hard-to-expressed genes, at least one has clear uORFs that might have limited the translation. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Plants 10.1038/s41477-022-01136-8
During my research journey, I have come across a handful of genes that are hard to express. Sometimes, even with a strong promoter and high transcript level, the protein level is negligible. We generally considered these proteins might be degrading very fast. But maybe not. A recent article by Wu et al. indicates translation of genes could be controlled by an upstream open reading frame (uORF). uORFs are upstream elements in the transcript that regulate the translation of about 30-50% of genes. The authors found that genes lacking uORF show leaky protein expression but the genes that have uORF don’t show this leaky expression. The authors observed the presence of uORF in a circadian-regulated gene TOC1. They mutated the uORF of TOC1 and observed that the mutated version produced more protein per mRNA than TOC1 harboring the normal uORF, indicating this uORF reduces protein expression. The authors also observed that the lack of proper uORF in Arabidopsis cells leads to loss of rhythmicity of TOC1 protein level, and that the uORF decreases cell-to-cell variability in TOC1 rhythmicity; that by buffering the expression level, noise is also reduced. They created a delightful animation that explains how this works. Overall, uORF elements seem to check undesired protein expression. Looking at the transcript of my hard-to-expressed genes, at least one has clear uORFs that might have limited the translation. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Plants 10.1038/s41477-022-01136-8
KIL1 terminates fertility in maize by controlling silk senescence
 Each plant species has a defined window of time during which flowers can be pollinated in order to trigger fruit or seed development. After this time, unpollinated flowers undergo a form of senescence culminating in programmed cell death (PCD). A detailed understanding of this cell death process is thus important to generate new varieties of agronomic crops with optimized yields. Here Šimášková et al. show that the transcription factor KIL1 is an important regulator of silk base senescence and PCD in maize. The group carried out RNA-sequencing on flowers at various stages of senescence, and found that some of the most highly upregulated genes during senescence were 5 orthologs of the Arabidopsis NAC transcription factor KIR1. These genes, named KIL1-5, were able to induce cell death when overexpressed in both maize protoplasts and Nicotiana benthamiana leaves. Similarly KIL1 overexpression in maize plants promoted early senescence while kil1 knock-out plants displayed delayed senescence and PCD. Crucially kil1 plants also exhibited a longer window of fertility and greater kernel set than wild type when pollinated late in the window, suggesting a possible method increasing yield in this vital crop. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koac151
Each plant species has a defined window of time during which flowers can be pollinated in order to trigger fruit or seed development. After this time, unpollinated flowers undergo a form of senescence culminating in programmed cell death (PCD). A detailed understanding of this cell death process is thus important to generate new varieties of agronomic crops with optimized yields. Here Šimášková et al. show that the transcription factor KIL1 is an important regulator of silk base senescence and PCD in maize. The group carried out RNA-sequencing on flowers at various stages of senescence, and found that some of the most highly upregulated genes during senescence were 5 orthologs of the Arabidopsis NAC transcription factor KIR1. These genes, named KIL1-5, were able to induce cell death when overexpressed in both maize protoplasts and Nicotiana benthamiana leaves. Similarly KIL1 overexpression in maize plants promoted early senescence while kil1 knock-out plants displayed delayed senescence and PCD. Crucially kil1 plants also exhibited a longer window of fertility and greater kernel set than wild type when pollinated late in the window, suggesting a possible method increasing yield in this vital crop. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koac151
Biofortified tomatoes provide a new route to vitamin D sufficiency
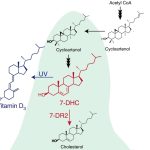
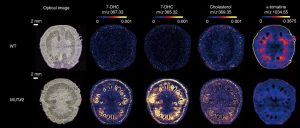 Vitamin D deficiency in humans is correlated with malfunction of the immune system and inflammation together with cancer, Parkinson disease, depression, neurocognitive decline, dementia, and severe COVID-19 infection. In humans, exposure of the skin to UV light promotes the production of Vitamin D from a precursor, 7-dehydrocholesterol (7-DHC), also known as provitamin D3, but most vitamin D has a dietary source. The best natural sources of vitamin D are fish and dairy products as the amount found in plants is generally low, which can be a problem for the vegan and vegetarian people. Provitamin D3 (7-DHC) is synthesized in tomato leaves through the steroidal glycoalkaloid (SGA) synthesis. It has been observed that there is a duplicated pathway in Solanaceous species for SGA synthesis. In this work, Li et al. engineer one of the SGA biosynthesis pathways for 7-DHC accumulation in tomato fruit and leaves. Using CRISPR/Cas9, the authors blocked 7-dehydrocholesterol reductase (Sl7-DR2) in order to avoid the conversion of 7-DHC to cholesterol. Sliced fruit and leaves with accumulated 7-DHC were exposed to UVB light resulting in high yields of vitamin D3. The loss of Sl7-DR2 activity had no effect on the growth, development or yield of tomato lines. This work suggests the use of tomato as a plant-based sustainable source of vitamin D3. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-022-01154-6
Vitamin D deficiency in humans is correlated with malfunction of the immune system and inflammation together with cancer, Parkinson disease, depression, neurocognitive decline, dementia, and severe COVID-19 infection. In humans, exposure of the skin to UV light promotes the production of Vitamin D from a precursor, 7-dehydrocholesterol (7-DHC), also known as provitamin D3, but most vitamin D has a dietary source. The best natural sources of vitamin D are fish and dairy products as the amount found in plants is generally low, which can be a problem for the vegan and vegetarian people. Provitamin D3 (7-DHC) is synthesized in tomato leaves through the steroidal glycoalkaloid (SGA) synthesis. It has been observed that there is a duplicated pathway in Solanaceous species for SGA synthesis. In this work, Li et al. engineer one of the SGA biosynthesis pathways for 7-DHC accumulation in tomato fruit and leaves. Using CRISPR/Cas9, the authors blocked 7-dehydrocholesterol reductase (Sl7-DR2) in order to avoid the conversion of 7-DHC to cholesterol. Sliced fruit and leaves with accumulated 7-DHC were exposed to UVB light resulting in high yields of vitamin D3. The loss of Sl7-DR2 activity had no effect on the growth, development or yield of tomato lines. This work suggests the use of tomato as a plant-based sustainable source of vitamin D3. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-022-01154-6