Plant Science Research Weekly: July 5th
Critical residues for carotenoid biosynthesis by phytoene synthase
 Carotenoids are diverse structures that contribute to photosynthetic light harvesting and serve as pigments, photoprotectants, and precursors for vitamin A and signalling molecules. Phytoene synthase is the first committed enzyme for carotenoid synthesis, and in tomato it is encoded by two genes, PSY1 and PSY2. When compared at similar protein levels, PSY1 expression results in much lower accumulation of carotenoids than PSY2, suggesting that it is a less efficient enzyme, although modelling studies suggest that both enzymes are structurally similar. To identify the cause of the different activities, Cao et al. identified several residues as potentially key. Through site-directed mutagenesis, they identified a pair of adjacent aromatic residues that are critical for high levels of activity. These high-activity associated residues are conserved in most other plants. Interestingly, PSY1 is the main enzyme expressed in tomato fruit, where high levels of transcription compensate for its lower relative activity. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00384
Carotenoids are diverse structures that contribute to photosynthetic light harvesting and serve as pigments, photoprotectants, and precursors for vitamin A and signalling molecules. Phytoene synthase is the first committed enzyme for carotenoid synthesis, and in tomato it is encoded by two genes, PSY1 and PSY2. When compared at similar protein levels, PSY1 expression results in much lower accumulation of carotenoids than PSY2, suggesting that it is a less efficient enzyme, although modelling studies suggest that both enzymes are structurally similar. To identify the cause of the different activities, Cao et al. identified several residues as potentially key. Through site-directed mutagenesis, they identified a pair of adjacent aromatic residues that are critical for high levels of activity. These high-activity associated residues are conserved in most other plants. Interestingly, PSY1 is the main enzyme expressed in tomato fruit, where high levels of transcription compensate for its lower relative activity. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.19.00384
Very long chain fatty acids (VLCFA) are involved in lateral root formation
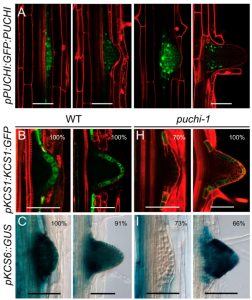 Root branching determines the spatial organization of the root and its interaction with the soil. The emergence of lateral roots (LR) is controlled by a complex regulatory network, involving genetic, hormonal and mechanical factors. In this study, Trinh et al. got new insights about PUCHI, a transcription factor previously shown to control cell proliferation during lateral root primordium (LRP) formation. Through co-expression analysis using time-course transcriptomic data of LRP organogenesis, the expression of many genes encoding enzymes involved in very long chain fatty acid (VLCFA; fatty acids with 20 or more carbons) biosynthesis was correlated with PUCHI expression. VLCFA are regulators of cell proliferation and important for membrane dynamics; the content and distribution of this kind of lipids can alter hormone signaling and affect the mechanical properties of the cell wall. The authors studied the expression patterns of the VLCFA biosynthesis genes in emerging LR. The density of emerged LR was not different between puchi-1 and WT, but the puchi-1 mutant has three times more LRP. With these data, they conclude PUCHI represses LRP initiation and the spacing between LRP. Other VLCFA mutants, kcs-5 and the cer10-2, had similar but less severe root branching defects. The authors also found that the control of VLCFA biosynthesis pathway by PUCHI controls cell proliferation in both lateral root development and callus formation. (Summary by Humberto Herrera-Ubaldo). Proc. Natl. Acad. Sci. USA
Root branching determines the spatial organization of the root and its interaction with the soil. The emergence of lateral roots (LR) is controlled by a complex regulatory network, involving genetic, hormonal and mechanical factors. In this study, Trinh et al. got new insights about PUCHI, a transcription factor previously shown to control cell proliferation during lateral root primordium (LRP) formation. Through co-expression analysis using time-course transcriptomic data of LRP organogenesis, the expression of many genes encoding enzymes involved in very long chain fatty acid (VLCFA; fatty acids with 20 or more carbons) biosynthesis was correlated with PUCHI expression. VLCFA are regulators of cell proliferation and important for membrane dynamics; the content and distribution of this kind of lipids can alter hormone signaling and affect the mechanical properties of the cell wall. The authors studied the expression patterns of the VLCFA biosynthesis genes in emerging LR. The density of emerged LR was not different between puchi-1 and WT, but the puchi-1 mutant has three times more LRP. With these data, they conclude PUCHI represses LRP initiation and the spacing between LRP. Other VLCFA mutants, kcs-5 and the cer10-2, had similar but less severe root branching defects. The authors also found that the control of VLCFA biosynthesis pathway by PUCHI controls cell proliferation in both lateral root development and callus formation. (Summary by Humberto Herrera-Ubaldo). Proc. Natl. Acad. Sci. USA
Cell cycle dependent regulation and function of ARGONAUTE1 in plants ($)
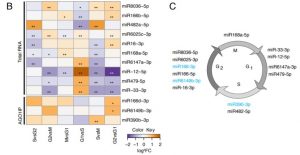 Regulation of gene expression at transcriptional, translational and post-translation levels is crucial for proper plant growth. Post-transcriptional gene regulation by small RNAs like microRNA (miRNA), siRNA and phasi-RNAs is necessary for meristem maintenance in plants. Unlike in the mammalian system, the role of small RNAs during the cell cycle in meristematic tissue is unknown. In this paper, Trolet et al. generated a large scale dataset that shines more light on small RNAs and specific regulators associated with ARGONAUTE1, which is involved in small RNA biogenesis. The loss of function ago1 mutants have altered meristem maintenance, and modulating AGO1 degradation during different phases of cell cycle causes changes in cell division at the Arabidopsis root meristem. Tobacco BY-2 cells were used to determine differentially expressed protein-coding mRNAs, small RNAs and cleaved target mRNAs at different stages of cell cycle. The miRNAs and differentially expressed genes that were identified from this study need to be further investigated for their roles in the cell cycle. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00069.
Regulation of gene expression at transcriptional, translational and post-translation levels is crucial for proper plant growth. Post-transcriptional gene regulation by small RNAs like microRNA (miRNA), siRNA and phasi-RNAs is necessary for meristem maintenance in plants. Unlike in the mammalian system, the role of small RNAs during the cell cycle in meristematic tissue is unknown. In this paper, Trolet et al. generated a large scale dataset that shines more light on small RNAs and specific regulators associated with ARGONAUTE1, which is involved in small RNA biogenesis. The loss of function ago1 mutants have altered meristem maintenance, and modulating AGO1 degradation during different phases of cell cycle causes changes in cell division at the Arabidopsis root meristem. Tobacco BY-2 cells were used to determine differentially expressed protein-coding mRNAs, small RNAs and cleaved target mRNAs at different stages of cell cycle. The miRNAs and differentially expressed genes that were identified from this study need to be further investigated for their roles in the cell cycle. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.19.00069.
A signal amplifier that regulates soybean root growth in response to salt stress
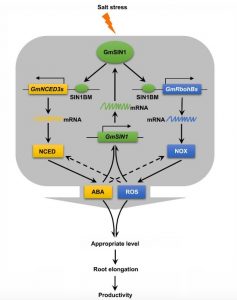
The balance between environmental stress tolerance and plant growth is an important research topic. Plant responses vary among species and varieties, but also with the growth environment. Soil, water, and environmental factors interact to affect plant growth responses to soil salinity. To engineer salt-tolerant crop varieties, it is crucial to identify the key components of the salt-tolerance network. Li et al. identified a possible candidate in soybean (Glycine max), a salinity-induced NAC transcription factor encoded by SALT INDUCED NAC 1 (GmSIN1). Upregulation of GmSIN1 depends on ABA and ROS. In response to salinity, GmSIN1 expression improves root growth and salt tolerance by promoting ABA accumulation and ROS generation, by up-regulating GmNCED3s and GmRbohBs. Based on this, GmSIN1, GmNCED3s and GmRbohBs collaborate to rapidly amplify the initial salt stress signal in a positive feed-forward system. These findings highlight that the modulation of ABA and ROS contents enhances soybean salt tolerance. (Summarized by Francesca Resentini) Plant Cell 10.1105/tpc.18.00662
RIN4/NOI fragments utilize different N-end rules of degradation to fight off pathogens
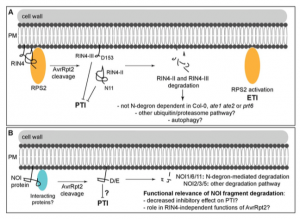 To fight off invading pathogens, plants such as Arabidopsis are intrinsically programmed with a subset of defense responses often known as pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). AvrRpt2, an effector secreted by the pathogen Pseudomonas syringe, is a protease that can cleave several central regulators of immune response in plants. The nitrate-induced domain (NOI) protein family in plants, release fragments generated by bacterial AvrRpt2 cleavage that can trigger host immune responses. It is thought that most of NOI protein fragments are destabilized by a N-degron pathway. However, recently Goslin et al. found an exception in a NOI protein called RPM1-INTERACTING PROTEIN4 (RIN4), whose destabilization of AvrRpt2 is independent of the N-degron pathway. This study advances our current understanding of the role of N-degron mediated degradation of NOI fragments in the context of the AvrRpt2 diversity. (Summarized by Nanxun Qin) Plant Physiology 10.1104/pp.19.00251
To fight off invading pathogens, plants such as Arabidopsis are intrinsically programmed with a subset of defense responses often known as pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). AvrRpt2, an effector secreted by the pathogen Pseudomonas syringe, is a protease that can cleave several central regulators of immune response in plants. The nitrate-induced domain (NOI) protein family in plants, release fragments generated by bacterial AvrRpt2 cleavage that can trigger host immune responses. It is thought that most of NOI protein fragments are destabilized by a N-degron pathway. However, recently Goslin et al. found an exception in a NOI protein called RPM1-INTERACTING PROTEIN4 (RIN4), whose destabilization of AvrRpt2 is independent of the N-degron pathway. This study advances our current understanding of the role of N-degron mediated degradation of NOI fragments in the context of the AvrRpt2 diversity. (Summarized by Nanxun Qin) Plant Physiology 10.1104/pp.19.00251
PEN3 and PDR12 secrete camalexin to the apoplast to limit pathogen growth in Arabidopsis
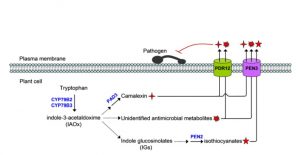 Phytoalexins are important antimicrobial compounds that plants synthesize to fend off invading pathogens. In the Brassica family, the tryptophan (trp)-derived phytoalexin ‘camalexin’ provides broad-spectrum resistance against bacterial, fungal, and oomycete pathogens. The regulation of camalexin biosynthesis during pathogen attack is well described, however a mechanistic understanding of its transport towards invading pathogens was previously unresolved. In a new study, He et al. leveraged the role of the PDR (Pleiotropic Drug Resistance) family transporter PEN3 (Penetration3), previously implicated in the secretion of an unknown complement of phytoalexins, to identify transporters involved in the secretion of camalexin in Arabidopsis. The authors first profiled the expression of each PDR family member during infection with the fungal pathogen Botrytis cinerea using qRT-PCR and GUS reporter analyses, revealing a strong upregulation of PEN3 (PDR8) and PDR12. Phenotypic analyses demonstrated a redundant role for PEN3 and PDR12 in providing resistance to B. cinerea, such that pen3/pdr12 double mutants were drastically more susceptible to infection than either single mutant or wild-type controls, and Botrytis-infected pen3/pdr12 double mutants contain abnormally high intracellular levels of camalexin due to significant defects in its secretion into the apoplast. Consistent with these results, pen3/pdr12 seedlings exhibited an enhanced sensitivity to exogenous camalexin treatment. Further biochemical analyses supported a role for PEN3 and PDR12 in transporting an unknown set of additional trp-derived metabolites and supported the well-established role for PEN3 (but not PDR12) in the transport of indole-glucosinolates (IGs). This work furthers our current understanding of phytoalexin transport during pathogen infection in Arabidopsis and provides novel insight into the role of PDR family transporters in plants. (Summary by Phil Carella) Plant Cell 10.1105/tpc.19.00239
Phytoalexins are important antimicrobial compounds that plants synthesize to fend off invading pathogens. In the Brassica family, the tryptophan (trp)-derived phytoalexin ‘camalexin’ provides broad-spectrum resistance against bacterial, fungal, and oomycete pathogens. The regulation of camalexin biosynthesis during pathogen attack is well described, however a mechanistic understanding of its transport towards invading pathogens was previously unresolved. In a new study, He et al. leveraged the role of the PDR (Pleiotropic Drug Resistance) family transporter PEN3 (Penetration3), previously implicated in the secretion of an unknown complement of phytoalexins, to identify transporters involved in the secretion of camalexin in Arabidopsis. The authors first profiled the expression of each PDR family member during infection with the fungal pathogen Botrytis cinerea using qRT-PCR and GUS reporter analyses, revealing a strong upregulation of PEN3 (PDR8) and PDR12. Phenotypic analyses demonstrated a redundant role for PEN3 and PDR12 in providing resistance to B. cinerea, such that pen3/pdr12 double mutants were drastically more susceptible to infection than either single mutant or wild-type controls, and Botrytis-infected pen3/pdr12 double mutants contain abnormally high intracellular levels of camalexin due to significant defects in its secretion into the apoplast. Consistent with these results, pen3/pdr12 seedlings exhibited an enhanced sensitivity to exogenous camalexin treatment. Further biochemical analyses supported a role for PEN3 and PDR12 in transporting an unknown set of additional trp-derived metabolites and supported the well-established role for PEN3 (but not PDR12) in the transport of indole-glucosinolates (IGs). This work furthers our current understanding of phytoalexin transport during pathogen infection in Arabidopsis and provides novel insight into the role of PDR family transporters in plants. (Summary by Phil Carella) Plant Cell 10.1105/tpc.19.00239
Functional traits and phenotypic plasticity modulate species coexistence across contrasting climatic conditions
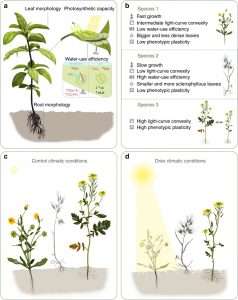 Understanding mechanisms for assembly of plant communities involves estimating different interactions among coexisting species, and how environmental change might affect those interactions. Peréz-Ramos et al. address how functional traits and their plasticity relate to the mechanisms that allow species to coexist. They studied ten annual species under two contrasting water availability conditions (control and drought). Traits related to plant physiology, morphology, phenology and reproductive ability were measured. For quantification of niche and fitness differences, mathematical demographic models of community assembly were used. Uni- and multidimensional associations were found highlighting for example the importance of light- and water-use efficiency. The plasticity of such traits modulates species coexistence at contrasting environments by stabilizing niche differences and by generating competitive trade-offs between species. (Summary by Cecilia Vasquez-Robinet) Nature Comms. 10.1038/s41467-019-10453-0.
Understanding mechanisms for assembly of plant communities involves estimating different interactions among coexisting species, and how environmental change might affect those interactions. Peréz-Ramos et al. address how functional traits and their plasticity relate to the mechanisms that allow species to coexist. They studied ten annual species under two contrasting water availability conditions (control and drought). Traits related to plant physiology, morphology, phenology and reproductive ability were measured. For quantification of niche and fitness differences, mathematical demographic models of community assembly were used. Uni- and multidimensional associations were found highlighting for example the importance of light- and water-use efficiency. The plasticity of such traits modulates species coexistence at contrasting environments by stabilizing niche differences and by generating competitive trade-offs between species. (Summary by Cecilia Vasquez-Robinet) Nature Comms. 10.1038/s41467-019-10453-0.