Plant Science Research Weekly: July 30, 2021
Review: Convergent evolution of gene regulatory networks underlying plant adaptations
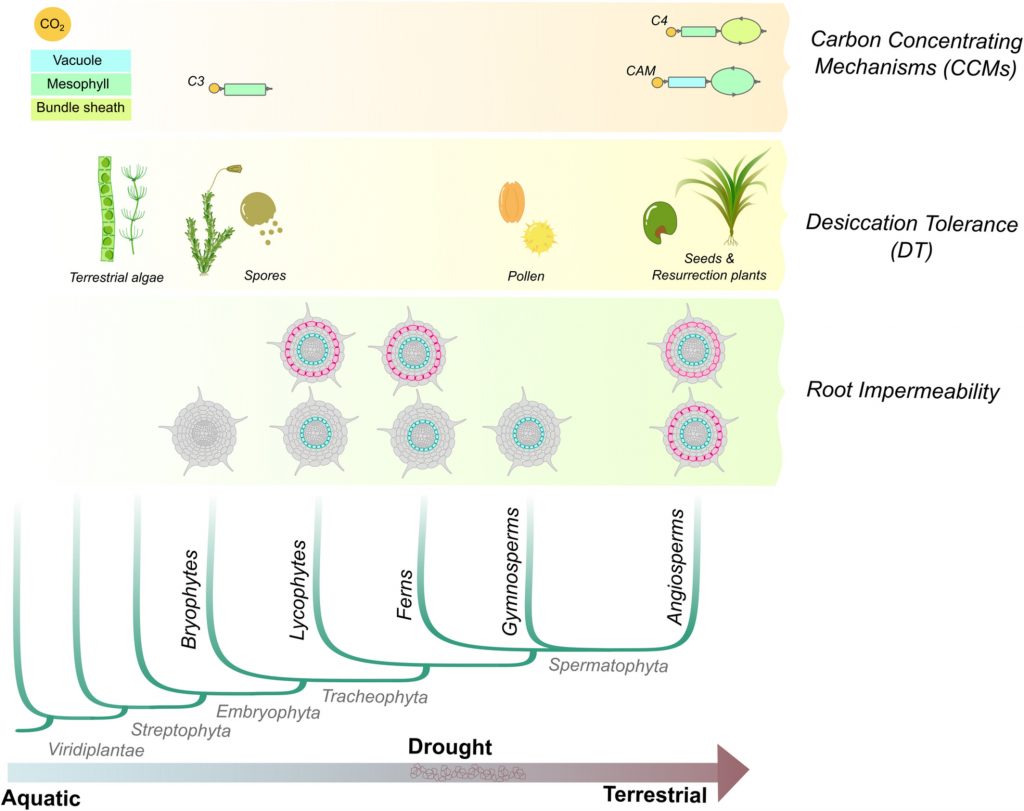 The transition from aquatic to terrestrial habitats exposed plants to low water availability, high light, radiation, and other environmental challenges. To overcome these challenges, plants developed morpho-physiological adaptations to tolerate dry environments and make photosynthesis more efficient. A recent work by M. Artur and K. Kajala reviewed how the evolution of Gene Regulatory Networks has shaped plant adaptations to dry environments. Carbon concentration, desiccation tolerance, and the production of extracellular biopolymers are common mechanisms in land plants that appeared multiple times during evolution. Desiccation tolerance is present in many plants’ reproductive structures (spores, pollen, seeds), but some of these regulatory pathways were re-directed to vegetative tissues in some species. Analysis of desiccation-tolerant species from different phylogenetic groups has revealed common mechanisms to achieve this ability to survive after extreme dehydration. In the synthesis of biopolymers that act as barriers to prevent water loss (i.e., cutin and suberin), the production of precursor molecules was possible before the evolution of land plants; however, the rest of the pathways may have evolved independently in the different taxa. This work highlights the importance of genomic and transcriptomic datasets available from plants of distinct clades to compare and trace the evolutionary history of critical regulators across the tree of life. (Summary by Humberto Herrera-Ubaldo @herrera_h) Plant Cell Envion. 10.1111/pce.14143
The transition from aquatic to terrestrial habitats exposed plants to low water availability, high light, radiation, and other environmental challenges. To overcome these challenges, plants developed morpho-physiological adaptations to tolerate dry environments and make photosynthesis more efficient. A recent work by M. Artur and K. Kajala reviewed how the evolution of Gene Regulatory Networks has shaped plant adaptations to dry environments. Carbon concentration, desiccation tolerance, and the production of extracellular biopolymers are common mechanisms in land plants that appeared multiple times during evolution. Desiccation tolerance is present in many plants’ reproductive structures (spores, pollen, seeds), but some of these regulatory pathways were re-directed to vegetative tissues in some species. Analysis of desiccation-tolerant species from different phylogenetic groups has revealed common mechanisms to achieve this ability to survive after extreme dehydration. In the synthesis of biopolymers that act as barriers to prevent water loss (i.e., cutin and suberin), the production of precursor molecules was possible before the evolution of land plants; however, the rest of the pathways may have evolved independently in the different taxa. This work highlights the importance of genomic and transcriptomic datasets available from plants of distinct clades to compare and trace the evolutionary history of critical regulators across the tree of life. (Summary by Humberto Herrera-Ubaldo @herrera_h) Plant Cell Envion. 10.1111/pce.14143
Review: Fleshy structures associated with ovule protection and seed dispersal in gymnosperms
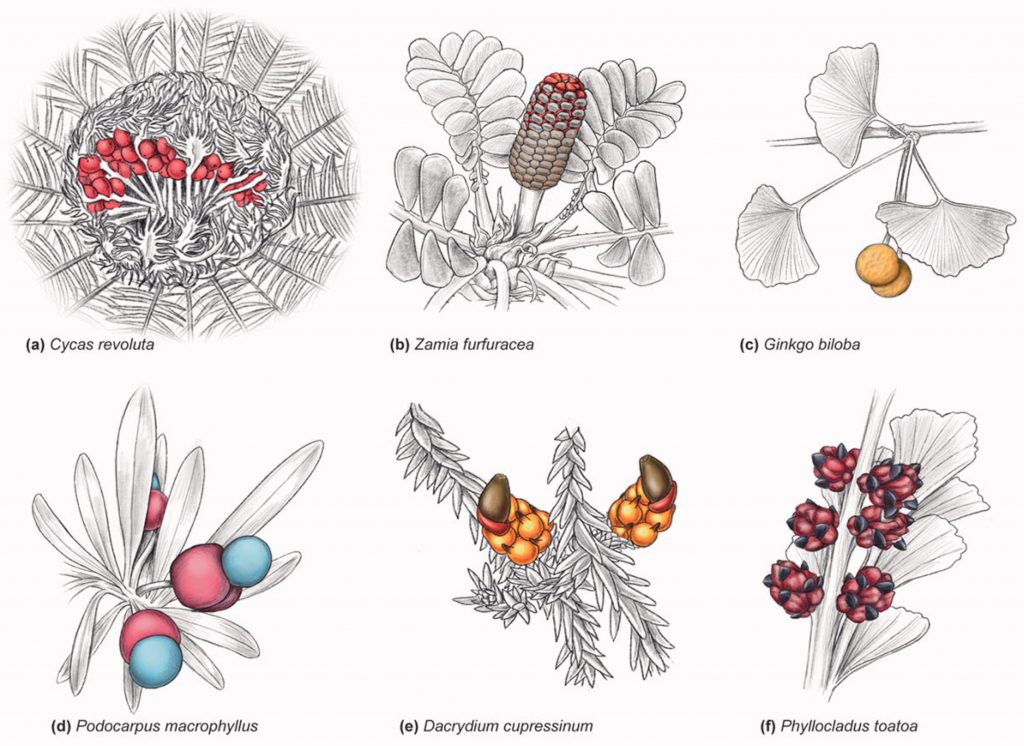 While traditionally known for having their ovules exposed to the environment, all gymnosperm lineages have species with fleshy seed-associated tissues. Here, Nigris and colleagues provide an overview of the vast diversity of fruit-like structures present in gymnosperms and the hypothesis available to explain their origin. Fleshy tissues have evolved multiple independent times through the evolution of this group, even in quite distant geological times. Moreover, they have different developmental origins depending on the group: from the ovule integument in Cycas and Gingko, passing by the ovule funiculus in Taxus, to various cone structures in Podocarpaceae and Ephedra. Given this diversity, various hypotheses have been proposed to explain the origin of fruit-like structures in gymnosperms, but most of them remain to be formally tested. Interestingly, recent studies have shown that the development of these structures shares some of the pathways found in angiosperm fruit development, such as the master regulation of MADS-box genes and the softening of tissues mediated by ethylene. Therefore, this review provides an exciting summary of the diversity of fleshy structures associated with gymnosperms seeds and a promising research agenda to unravel their origin. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Crit. Rev. Plant Sci. 10.1080/07352689.2021.1938397
While traditionally known for having their ovules exposed to the environment, all gymnosperm lineages have species with fleshy seed-associated tissues. Here, Nigris and colleagues provide an overview of the vast diversity of fruit-like structures present in gymnosperms and the hypothesis available to explain their origin. Fleshy tissues have evolved multiple independent times through the evolution of this group, even in quite distant geological times. Moreover, they have different developmental origins depending on the group: from the ovule integument in Cycas and Gingko, passing by the ovule funiculus in Taxus, to various cone structures in Podocarpaceae and Ephedra. Given this diversity, various hypotheses have been proposed to explain the origin of fruit-like structures in gymnosperms, but most of them remain to be formally tested. Interestingly, recent studies have shown that the development of these structures shares some of the pathways found in angiosperm fruit development, such as the master regulation of MADS-box genes and the softening of tissues mediated by ethylene. Therefore, this review provides an exciting summary of the diversity of fleshy structures associated with gymnosperms seeds and a promising research agenda to unravel their origin. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Crit. Rev. Plant Sci. 10.1080/07352689.2021.1938397
Opinion: Plants have neither synapses nor a nervous system
 The question “Are plants intelligent?” is raised regularly, with answer often “Well, it depends how you define intelligence.” Another interesting question is whether or not plants have cellular structures that are analogous to those that make up the animal nervous system. Robinson and Draguhn argue that they do not, in this carefully constructed Opinion article. This is an intriguing article that could be compelling to students and useful for teaching. The authors, a plant cell biologist and a neuroscientist, compare and contrast features of the animal nervous system (synapses, neurons) to structures in plants (plasmodesmata, phloem system), and conclude that although plants “certainly do have complex cell contacts and signaling mechanisms, none of these structures provides a basis for neuronal-like synaptic transmission.” (Summary by Mary Williams @PlantTeaching) J. Plant Physiol. 10.1016/j.jplph.2021.153467
The question “Are plants intelligent?” is raised regularly, with answer often “Well, it depends how you define intelligence.” Another interesting question is whether or not plants have cellular structures that are analogous to those that make up the animal nervous system. Robinson and Draguhn argue that they do not, in this carefully constructed Opinion article. This is an intriguing article that could be compelling to students and useful for teaching. The authors, a plant cell biologist and a neuroscientist, compare and contrast features of the animal nervous system (synapses, neurons) to structures in plants (plasmodesmata, phloem system), and conclude that although plants “certainly do have complex cell contacts and signaling mechanisms, none of these structures provides a basis for neuronal-like synaptic transmission.” (Summary by Mary Williams @PlantTeaching) J. Plant Physiol. 10.1016/j.jplph.2021.153467
Nurse cell–derived small RNAs define paternal epigenetic inheritance in Arabidopsis
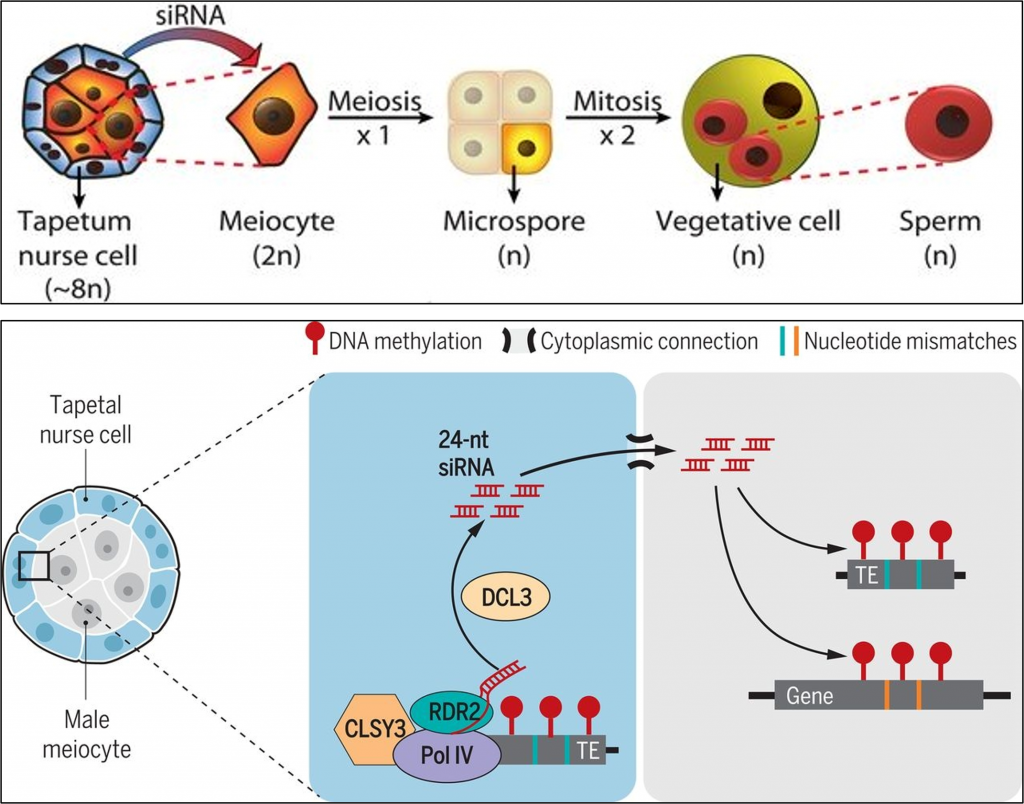 New mechanisms for the production and function of small RNAs continue to be discovered. Previous studies have shown a role for small RNAs in methylation reprogramming. Recently, Long et al. found that the epigenetic reprogramming that takes place in the male reproductive tissues also involves mobile small RNAs. They found that small RNAs are produced by RNA-dependent DNA polymerase in tapetal nurse cells of the anther – these cells are diploid, somatic cells, but they surround and support the development of the male meiocytes that will form the sperm cells. The meiocytes undergo extensive DNA methylation reprogramming, but they do not themselves produce the 24-nt siRNAs that mediate it. The authors found that these siRNA are produced in the nurse cells and move into the meiocytes through plasmodesmata. Interestingly, the siRNAs are transcribed in the nurse cells from transposons, which they target in these cells, whereas within the meiocytes they define paternal epigenetic inheritance by also targeting genes with imperfect sequence matches. This so-called trans-homologue methylation of protein-coding genes does not occur in the nurse cells, it is meiocyte-specific. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abh0556 This article is also available as a preprint https://www.biorxiv.org/content/10.1101/2021.01.25.428150v1 See also commentary by Rebecca Mosher 10.1126/science.abj5020
New mechanisms for the production and function of small RNAs continue to be discovered. Previous studies have shown a role for small RNAs in methylation reprogramming. Recently, Long et al. found that the epigenetic reprogramming that takes place in the male reproductive tissues also involves mobile small RNAs. They found that small RNAs are produced by RNA-dependent DNA polymerase in tapetal nurse cells of the anther – these cells are diploid, somatic cells, but they surround and support the development of the male meiocytes that will form the sperm cells. The meiocytes undergo extensive DNA methylation reprogramming, but they do not themselves produce the 24-nt siRNAs that mediate it. The authors found that these siRNA are produced in the nurse cells and move into the meiocytes through plasmodesmata. Interestingly, the siRNAs are transcribed in the nurse cells from transposons, which they target in these cells, whereas within the meiocytes they define paternal epigenetic inheritance by also targeting genes with imperfect sequence matches. This so-called trans-homologue methylation of protein-coding genes does not occur in the nurse cells, it is meiocyte-specific. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abh0556 This article is also available as a preprint https://www.biorxiv.org/content/10.1101/2021.01.25.428150v1 See also commentary by Rebecca Mosher 10.1126/science.abj5020
Complex networks of prion-like proteins reveal cross talk between stress and memory pathways in plants
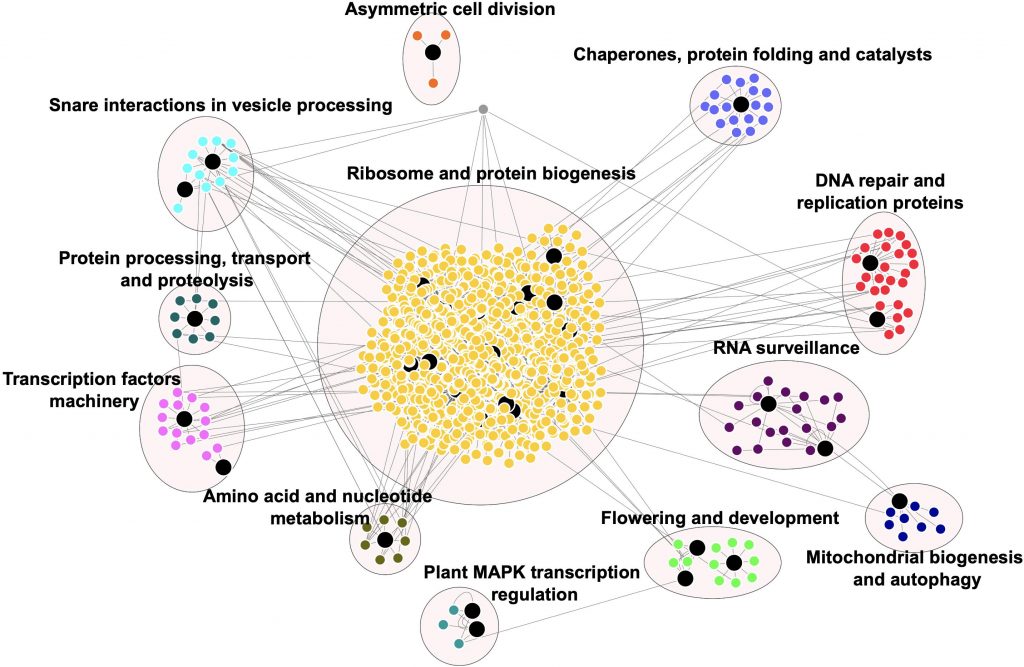 Prions were first identified as the infectious protein agents causative of Creutzfeldt-Jakob disease or bovine spongiform encephalopathy (mad cow disease); when individuals eat something containing these prion proteins, the prion proteins induce conformational changes in their brains leading to neurological damage. Prions are more generally and less scarily now known as proteins that can form different structures and initiate reversible biomolecular condensates or liquid-liquid phase separation. Previous work has shown prion-like proteins as having a role in stress and memory in yeast and Arabidopsis. Here, Garai et al. investigate the plant “prionome” across 39 genomes, examining expression, gene regulatory networks, and interactomes of plant prion-like proteins. Their findings provide support for a role in these plant prion-like proteins in stress and memory. (Summary by Mary Williams @PlantTeaching) Frontiers Plant Sci. 10.3389/fpls.2021.707286
Prions were first identified as the infectious protein agents causative of Creutzfeldt-Jakob disease or bovine spongiform encephalopathy (mad cow disease); when individuals eat something containing these prion proteins, the prion proteins induce conformational changes in their brains leading to neurological damage. Prions are more generally and less scarily now known as proteins that can form different structures and initiate reversible biomolecular condensates or liquid-liquid phase separation. Previous work has shown prion-like proteins as having a role in stress and memory in yeast and Arabidopsis. Here, Garai et al. investigate the plant “prionome” across 39 genomes, examining expression, gene regulatory networks, and interactomes of plant prion-like proteins. Their findings provide support for a role in these plant prion-like proteins in stress and memory. (Summary by Mary Williams @PlantTeaching) Frontiers Plant Sci. 10.3389/fpls.2021.707286
Interested in writing for this series? Plantae Fellows applications are open now, deadline Aug 20, 2021. https://plantae.org/fellowsapp/.