Plant Science Research Weekly: July 26, 2024
Review: Stem cells of the vascular cambium
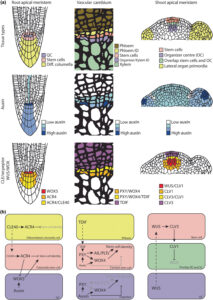 Vascular cambium cells are meristematic cells responsible for secondary growth. Lineage tracing studies in Arabidopsis and poplar show the presence of single bifacial stem cells in each radial cell file that produce xylem inwards and phloem cells outwards. In a recent review, Wybouw et al. discuss the concept of stem cells in vascular cambium. The review begins with a discussion of how secondary growth is mediated by phytohormones and peptide signaling. Next, they discuss three stem cell identities found within the vascular cambium: stem cells, organizer cells, and transit-amplifying cells. The stem cells are capable of continuously resupplying themselves through division. The organizer cells nonautonomously promote stem cell identity in the adjacent cells, Transit-amplifying cells are dividing cells of which daughter cells lose dividing capacity over time, although studying transit-amplifying cells is difficult due to low occurrence in Arabidopsis. Studying stem cells is also difficult due to challenges in positioning them in vascular cambium. While lineage studies can retrospectively inform the position of stem cells, more precise methods such as live imaging of cell division using confocal microscopy is not yet possible as cambium tissues are deep inside the plant. The authors hope that future studies can elaborate the exact roles of factors discussed and the possibility of translating these findings into other species besides Arabidopsis and poplar. (Summary by Kumanan N Govaichelvan, @NGKumanan) New Phtyol. 10.1111/nph.19897
Vascular cambium cells are meristematic cells responsible for secondary growth. Lineage tracing studies in Arabidopsis and poplar show the presence of single bifacial stem cells in each radial cell file that produce xylem inwards and phloem cells outwards. In a recent review, Wybouw et al. discuss the concept of stem cells in vascular cambium. The review begins with a discussion of how secondary growth is mediated by phytohormones and peptide signaling. Next, they discuss three stem cell identities found within the vascular cambium: stem cells, organizer cells, and transit-amplifying cells. The stem cells are capable of continuously resupplying themselves through division. The organizer cells nonautonomously promote stem cell identity in the adjacent cells, Transit-amplifying cells are dividing cells of which daughter cells lose dividing capacity over time, although studying transit-amplifying cells is difficult due to low occurrence in Arabidopsis. Studying stem cells is also difficult due to challenges in positioning them in vascular cambium. While lineage studies can retrospectively inform the position of stem cells, more precise methods such as live imaging of cell division using confocal microscopy is not yet possible as cambium tissues are deep inside the plant. The authors hope that future studies can elaborate the exact roles of factors discussed and the possibility of translating these findings into other species besides Arabidopsis and poplar. (Summary by Kumanan N Govaichelvan, @NGKumanan) New Phtyol. 10.1111/nph.19897
Review: Role of silicon in drought tolerance
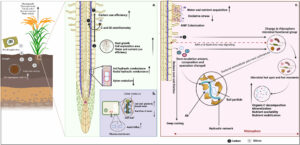 There is increasing evidence that silicon (Si) application to soils or leaves can promote plant growth through various means, including enhancing tolerance to abiotic and biotic stresses, but the mechanisms of this growth promotion are not always clear. Here, Bardhan et al. review how Si can promote drought tolerance, focusing on the soil-root interface. Starting with the soil, it has been shown that Si fertilization improves soil water retention, resulting in increased water availability to plants, and Si can also enhance nutrient availability in soil through effects on soil pH. Different plants accumulate different amounts of Si, and their responses to Si augmentation vary, but the collective list of positive benefits of Si is significant. For example, Si can interact with plant cell walls, for example by providing strength with a lower requirement for lignin, which frees up carbon for other functions, and strengthening xylem walls to prevent cavitation. Furthermore, the presence of Si can promote growth of root hairs and lateral roots, providing the infrastructure for water uptake. Some studies have also shown that Si indirectly enhances plant growth through effects on the plant microbiome. Although we still have gaps in our understanding of some of the ways that Si promotes drought tolerance, the evidence to support is use as a soil fertilizers is impressive. (Summary by Mary Williams @PlantTeaching) J Agron Crop Sci 10.1111/jac.12721
There is increasing evidence that silicon (Si) application to soils or leaves can promote plant growth through various means, including enhancing tolerance to abiotic and biotic stresses, but the mechanisms of this growth promotion are not always clear. Here, Bardhan et al. review how Si can promote drought tolerance, focusing on the soil-root interface. Starting with the soil, it has been shown that Si fertilization improves soil water retention, resulting in increased water availability to plants, and Si can also enhance nutrient availability in soil through effects on soil pH. Different plants accumulate different amounts of Si, and their responses to Si augmentation vary, but the collective list of positive benefits of Si is significant. For example, Si can interact with plant cell walls, for example by providing strength with a lower requirement for lignin, which frees up carbon for other functions, and strengthening xylem walls to prevent cavitation. Furthermore, the presence of Si can promote growth of root hairs and lateral roots, providing the infrastructure for water uptake. Some studies have also shown that Si indirectly enhances plant growth through effects on the plant microbiome. Although we still have gaps in our understanding of some of the ways that Si promotes drought tolerance, the evidence to support is use as a soil fertilizers is impressive. (Summary by Mary Williams @PlantTeaching) J Agron Crop Sci 10.1111/jac.12721
Transposase-assisted target-site integration for efficient plant genome engineering
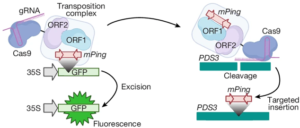 Genome editing stands at the forefront of scientific innovation, offering transformative possibilities to manipulate the genetic code of plants, animals, and humans. A critical bottleneck for its application in modern crop improvement is the low frequency and error-prone integration of foreign DNA into the plant genome. While the rapid development of CRISPR/Cas system, which functions like a pair of molecular “scissors” to cut the genome, allows site-specific changes to the DNA, current methods lack robust ways to add custom DNA accurately and efficiently at the cut sites. To address this, Liu and colleagues developed a “cut-and-paste” genome editing toolkit using Transposase-Assisted Target Site Integration (TATSI) technology. This method significantly improves the efficiency and accuracy of DNA integration. Plant transposable elements naturally function as molecular “glue” to insert DNA into the genome. When combined with CRISPR/Cas, this “scissors + glue” combination enables custom DNA integration at specific sites within the plant genome. This innovative system has proven effective in the model plant Arabidopsis and soybean, a rapidly growing crop model due to its critical role as a global source of protein and oil. This advancement in genome editing technology holds great promise for enhancing crop improvement and addressing global agricultural challenges. (Summary by Ching Chan @ntnuchanlab) Nature 10.1038/s41586-024-07613-8
Genome editing stands at the forefront of scientific innovation, offering transformative possibilities to manipulate the genetic code of plants, animals, and humans. A critical bottleneck for its application in modern crop improvement is the low frequency and error-prone integration of foreign DNA into the plant genome. While the rapid development of CRISPR/Cas system, which functions like a pair of molecular “scissors” to cut the genome, allows site-specific changes to the DNA, current methods lack robust ways to add custom DNA accurately and efficiently at the cut sites. To address this, Liu and colleagues developed a “cut-and-paste” genome editing toolkit using Transposase-Assisted Target Site Integration (TATSI) technology. This method significantly improves the efficiency and accuracy of DNA integration. Plant transposable elements naturally function as molecular “glue” to insert DNA into the genome. When combined with CRISPR/Cas, this “scissors + glue” combination enables custom DNA integration at specific sites within the plant genome. This innovative system has proven effective in the model plant Arabidopsis and soybean, a rapidly growing crop model due to its critical role as a global source of protein and oil. This advancement in genome editing technology holds great promise for enhancing crop improvement and addressing global agricultural challenges. (Summary by Ching Chan @ntnuchanlab) Nature 10.1038/s41586-024-07613-8
Identification of plant transcriptional activation domains
 Activation domains (AD) are parts of transcription factor proteins (TFs) that bind the transcriptional machinery (coactivator complexes) and lead to transcriptional activation. However, identifying ADs is challenging because they are often part of intrinsically disordered regions, which lack a defined 3D structure and conserved sequence motifs. In this paper, Morffy and colleagues adressed this challenge by combining large scale high throughput assays in yeast with the implementation of a neural network to identify and classify the ADs of plant TFs. To test the activity of ADs and identify their sequences, they created a library of 40aa long peptides spanning the whole coding sequence of 1918 TFs from Arabidopsis, and employed it in a fluorescence-based reporter system followed by sequencing. The experimental data were used to train a neural network that could identify ADs based on a set of predefined biochemical features derived from the amino acidic sequences. By extracting the features with the strongest effects from the model, the authors classified the ADs into six different subtypes, whose activity was then verified in planta. Finally, the authors applied the neuronal network to members of the Auxin Response Factor (ARFs) family across 117 angiosperms. They found that while the sequence similarity of ADs was low among orthologs, their location within the protein was conserved. This work provides unprecedented insights on the physico-chemical properties of TFs and, more broadly, on the mechanisms governing transcriptional activation. It emphasizes the role of positional and functional conservation, rather than sequence homology, in the evolution of TFs. (Summary by Carlo Pasini @Crl_Psn) Nature 10.1038/s41586-024-07707-3
Activation domains (AD) are parts of transcription factor proteins (TFs) that bind the transcriptional machinery (coactivator complexes) and lead to transcriptional activation. However, identifying ADs is challenging because they are often part of intrinsically disordered regions, which lack a defined 3D structure and conserved sequence motifs. In this paper, Morffy and colleagues adressed this challenge by combining large scale high throughput assays in yeast with the implementation of a neural network to identify and classify the ADs of plant TFs. To test the activity of ADs and identify their sequences, they created a library of 40aa long peptides spanning the whole coding sequence of 1918 TFs from Arabidopsis, and employed it in a fluorescence-based reporter system followed by sequencing. The experimental data were used to train a neural network that could identify ADs based on a set of predefined biochemical features derived from the amino acidic sequences. By extracting the features with the strongest effects from the model, the authors classified the ADs into six different subtypes, whose activity was then verified in planta. Finally, the authors applied the neuronal network to members of the Auxin Response Factor (ARFs) family across 117 angiosperms. They found that while the sequence similarity of ADs was low among orthologs, their location within the protein was conserved. This work provides unprecedented insights on the physico-chemical properties of TFs and, more broadly, on the mechanisms governing transcriptional activation. It emphasizes the role of positional and functional conservation, rather than sequence homology, in the evolution of TFs. (Summary by Carlo Pasini @Crl_Psn) Nature 10.1038/s41586-024-07707-3
CarboTag for live-imaging of plant cell walls
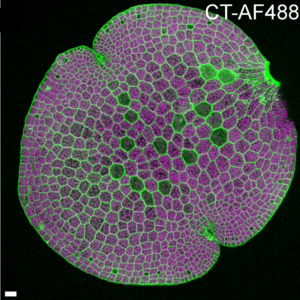 Recently, we have gained tremendous insights through live-cell imaging using fluorescent tags that bind specifically to various cellular components. Here, Besten et al. present a new set of cell wall-specific tags that bind specifically to cell walls and reveal selected wall properties. The authors developed a non-toxic, boron-based synthetic molecule, CarboTag, that binds to cell walls and can be linked to a variety of fluorophores. Because it does not require tissue fixation, it can be used for live-cell imaging. Furthermore, it works in a wide range of plants and even green and brown algae. Although it’s not completely clear what CarboTag binds to, evidence suggest it binds to hydrated carbohydrate hydrogels such as pectin, agar, and alginate. The authors also developed markers to visualize cell wall porosity, apoplastic pH, and ROS in cell walls. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.07.05.597952
Recently, we have gained tremendous insights through live-cell imaging using fluorescent tags that bind specifically to various cellular components. Here, Besten et al. present a new set of cell wall-specific tags that bind specifically to cell walls and reveal selected wall properties. The authors developed a non-toxic, boron-based synthetic molecule, CarboTag, that binds to cell walls and can be linked to a variety of fluorophores. Because it does not require tissue fixation, it can be used for live-cell imaging. Furthermore, it works in a wide range of plants and even green and brown algae. Although it’s not completely clear what CarboTag binds to, evidence suggest it binds to hydrated carbohydrate hydrogels such as pectin, agar, and alginate. The authors also developed markers to visualize cell wall porosity, apoplastic pH, and ROS in cell walls. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.07.05.597952
Characterization of pyrenoid-based CO2-concentrating mechanism in hornworts
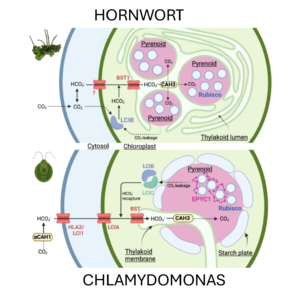 Pyrenoids are structures that concentrate carbon dioxide around Rubisco, most commonly studied in green algae such as Chlamydomonas. However, hornworts, one of the three types of bryophytes (along with liverworts and mosses) also have a pyrenoid-based CO2-concentrating mechanism (pCCM), unique among land plants. Here, Robison et al. characterize the pCCM in the model hornwort Anthoceros agrestis. The authors used electron microscopy and live-cell imaging to visualize the hornwort pCCM. They noted some differences from the pCCM of Chlamydomonas, including that hornworts have multiple pyrenoids in each chloroplast, in contrast to the single pyrenoid in Chlamydomonas. They further showed that the hornworts lack the starch sheath present in algal pyrenoids. They also used co-IP and fluorescent reporters to identify and characterize several pyrenoid-specific proteins, noting that many are present in the hornwort pCCM, but that the linker protein EPYC1 seems to be missing. As the authors report, characterizing the pCCM in hornworts provides new opportunities to engineer such structures in crops and other land plants. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.06.26.600872
Pyrenoids are structures that concentrate carbon dioxide around Rubisco, most commonly studied in green algae such as Chlamydomonas. However, hornworts, one of the three types of bryophytes (along with liverworts and mosses) also have a pyrenoid-based CO2-concentrating mechanism (pCCM), unique among land plants. Here, Robison et al. characterize the pCCM in the model hornwort Anthoceros agrestis. The authors used electron microscopy and live-cell imaging to visualize the hornwort pCCM. They noted some differences from the pCCM of Chlamydomonas, including that hornworts have multiple pyrenoids in each chloroplast, in contrast to the single pyrenoid in Chlamydomonas. They further showed that the hornworts lack the starch sheath present in algal pyrenoids. They also used co-IP and fluorescent reporters to identify and characterize several pyrenoid-specific proteins, noting that many are present in the hornwort pCCM, but that the linker protein EPYC1 seems to be missing. As the authors report, characterizing the pCCM in hornworts provides new opportunities to engineer such structures in crops and other land plants. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.06.26.600872
Tissue-specific interference reveals complex roles for auxin signalling in procambial cells during graft healing in Arabidopsis
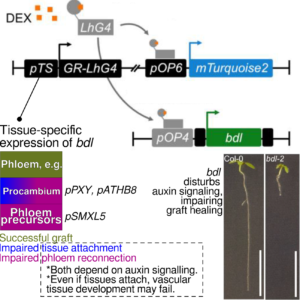 For hundreds of years, we have propagated plants clonally by grafting, i.e. the union of a scion and a rootstock to form a new plant. To be successful, a graft junction must heal: tissues must attach, and vascular bundles reconnect, in a process known to involve auxin. Here, Serivichyasw at et al. investigate the role of auxin signalling in different tissues during graft healing in Arabidopsis thaliana. The authors developed a platform for disturbing auxin signalling in an inducible, tissue-specific manner using promoter sequences of genes with cell type-specific expression patterns driving bdl, a nonfunctional BODENLOS/IAA12 allele that disturbs auxin signalling. Among the tested tissues, lines with procambium- and phloem precursor-localised expression of bdl saw the strongest losses in graft healing. Procambial cells expressing bdl caused failure of tissue attachment and phloem reconnection in grafts, much like what is seen for bdl-2 loss-of-function mutant lines. Although tissue attachment was successful in phloem precursor-specific bdl expression lines, phloem development in these grafts was impaired, suggesting the two processes are somewhat independent. Disturbing auxin signalling in the procambium also affected the critical callus formation stage of graft healing. Along with new insights into the biology of grafting, the expression platform developed herein could be used to better spatially resolve the role of auxin in other plant developmental processes. (Summary by John Vilasboa @vilasjohn) Plant Physiol. 10.1093/plphys/kiae257
For hundreds of years, we have propagated plants clonally by grafting, i.e. the union of a scion and a rootstock to form a new plant. To be successful, a graft junction must heal: tissues must attach, and vascular bundles reconnect, in a process known to involve auxin. Here, Serivichyasw at et al. investigate the role of auxin signalling in different tissues during graft healing in Arabidopsis thaliana. The authors developed a platform for disturbing auxin signalling in an inducible, tissue-specific manner using promoter sequences of genes with cell type-specific expression patterns driving bdl, a nonfunctional BODENLOS/IAA12 allele that disturbs auxin signalling. Among the tested tissues, lines with procambium- and phloem precursor-localised expression of bdl saw the strongest losses in graft healing. Procambial cells expressing bdl caused failure of tissue attachment and phloem reconnection in grafts, much like what is seen for bdl-2 loss-of-function mutant lines. Although tissue attachment was successful in phloem precursor-specific bdl expression lines, phloem development in these grafts was impaired, suggesting the two processes are somewhat independent. Disturbing auxin signalling in the procambium also affected the critical callus formation stage of graft healing. Along with new insights into the biology of grafting, the expression platform developed herein could be used to better spatially resolve the role of auxin in other plant developmental processes. (Summary by John Vilasboa @vilasjohn) Plant Physiol. 10.1093/plphys/kiae257
Rice tiller production regulated by sustained expression of FLORAL ORGAN NUMBER 1
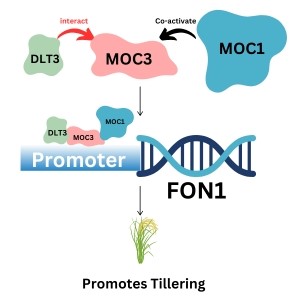 Number of tillers is one of the major determinants of yield in rice. MONOCULM1 (MOC1) and MOC3 have been reported to regulate tiller formation by promoting axillary bud formation and elongation of buds. MOC1 interacts with MOC3 and promotes the expression of FLORAL ORGAN NUMBER 1 (FON1), which increases the elongation of buds and ultimately promotes tillering. MOC3 can directly bind to the promoter of FON1, but MOC1 cannot directly bind to FON1 and acts as a co-activator of MOC3. Fan et al. discovered the function of a novel gene DWARF AND LESS TILLER ON CHROMOSOME 3 (DLT3), which interacts with MOC3, recruits MOC1, and binds to the promoter of FON1. They cloned DLT3 from a mutant developed from an EMS mutagenized population and performed Mutmap sequencing to find its candidate gene, whose function was validated by complementation assay and CRISPR genome editing. They found the potential interaction of DLT3 with MOC3 in a yeast two-hybrid screen and validated it through biomolecular fluorescence complementation. This discovery has added a new player, DLT3, to the previously known tiller regulating molecular circuit FON1-MOC3-MOC1. (Summary by Asif Ali @pbgasifkalas) Plant Physiol. 10.1093/plphys/kiae367
Number of tillers is one of the major determinants of yield in rice. MONOCULM1 (MOC1) and MOC3 have been reported to regulate tiller formation by promoting axillary bud formation and elongation of buds. MOC1 interacts with MOC3 and promotes the expression of FLORAL ORGAN NUMBER 1 (FON1), which increases the elongation of buds and ultimately promotes tillering. MOC3 can directly bind to the promoter of FON1, but MOC1 cannot directly bind to FON1 and acts as a co-activator of MOC3. Fan et al. discovered the function of a novel gene DWARF AND LESS TILLER ON CHROMOSOME 3 (DLT3), which interacts with MOC3, recruits MOC1, and binds to the promoter of FON1. They cloned DLT3 from a mutant developed from an EMS mutagenized population and performed Mutmap sequencing to find its candidate gene, whose function was validated by complementation assay and CRISPR genome editing. They found the potential interaction of DLT3 with MOC3 in a yeast two-hybrid screen and validated it through biomolecular fluorescence complementation. This discovery has added a new player, DLT3, to the previously known tiller regulating molecular circuit FON1-MOC3-MOC1. (Summary by Asif Ali @pbgasifkalas) Plant Physiol. 10.1093/plphys/kiae367
The white lupin trehalase gene regulates cluster root formation and function under phosphorus deficiency
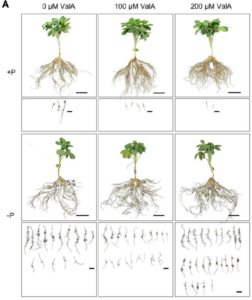 Phosphorus (P) is an essential nutrient for plant growth and development. Under P deficiency, white lupin develops cluster roots (CR), specialized root structures that enhance soil exploration and nutrient acquisition. While sugar signaling, particularly sucrose, has been shown to play a role in the establishment of CR, other CR-associated features such as the root exudation of citrate and malate are independent of sucrose. Therefore, a key sugar signaling component specific to CR function remains to be identified. Using a combination of pharmacological and genetic studies, Xia and colleagues demonstrated that trehalose plays a critical role in both CR establishment and organic acid metabolism. Specific inhibition of trehalase, the only enzyme that catalyzes trehalose catabolism, by validamycin A, promotes trehalose accumulation and CR formation under P deficiency conditions. Similarly, silencing the TREHALASE1 gene also promotes CR formation, while overexpressing the gene leads to an opposite effect. This study sheds light on the close association between trehalose metabolism and P starvation response in plants. (Summary by Ching Chan @ntnuchanlab) Plant Physiology 10.1093/plphys/kiae290
Phosphorus (P) is an essential nutrient for plant growth and development. Under P deficiency, white lupin develops cluster roots (CR), specialized root structures that enhance soil exploration and nutrient acquisition. While sugar signaling, particularly sucrose, has been shown to play a role in the establishment of CR, other CR-associated features such as the root exudation of citrate and malate are independent of sucrose. Therefore, a key sugar signaling component specific to CR function remains to be identified. Using a combination of pharmacological and genetic studies, Xia and colleagues demonstrated that trehalose plays a critical role in both CR establishment and organic acid metabolism. Specific inhibition of trehalase, the only enzyme that catalyzes trehalose catabolism, by validamycin A, promotes trehalose accumulation and CR formation under P deficiency conditions. Similarly, silencing the TREHALASE1 gene also promotes CR formation, while overexpressing the gene leads to an opposite effect. This study sheds light on the close association between trehalose metabolism and P starvation response in plants. (Summary by Ching Chan @ntnuchanlab) Plant Physiology 10.1093/plphys/kiae290
Pathogen cyclic lipopeptide virulence factors promote disease by inducing membrane leakage
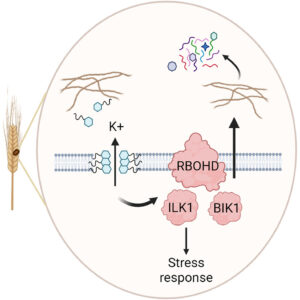 Pathogens deploy an array of molecules to create favorable conditions and promote infections in host plants. These molecules are well-known for suppressing host immunity, rendering them more susceptible. However, two recent reports suggest that some pathogen-produced virulence factors may also act independently of host immune suppression. Central to this are a class of molecules known as cyclic lipopeptides. In Cell Reports, Brauer et al. studied gramillin, secreted by the fungal pathogen Fusarium graminearum (causal agent of fusarium head blight), whereas in Nature Communications, Getzke and Wang et al. reported brassicapeptin A from the opportunistic bacterial pathogen Pseudomonas brassicacearum R401. Interestingly, both cyclic lipopeptides disrupt ionic balance in host plants through pore formation, with marginal suppression of host immunity. Specifically, gramillin induces reactive oxygen species (ROS) burst depending on host ILK1 and RBOHD genes but promotes disease independently of the major immune signaling pathways (such as jasmonic acid, salicylic acid, and pattern recognition receptors/PRRs). On the other hand, brassicapeptin A is salt-inducible and exhibits detrimental effects by interacting with salt in the environment, disturbing plant ion homeostasis. Together, these studies establish the multifaceted role of cyclic lipopeptides and their interactions with environmental stressors in pathogen success. (Summary by Arijit Mukherjee @ArijitM61745830) Cell Reports 10.1016/j.celrep.2024.114384, Nature Comms 10.1038/s41467-024-48517-5
Pathogens deploy an array of molecules to create favorable conditions and promote infections in host plants. These molecules are well-known for suppressing host immunity, rendering them more susceptible. However, two recent reports suggest that some pathogen-produced virulence factors may also act independently of host immune suppression. Central to this are a class of molecules known as cyclic lipopeptides. In Cell Reports, Brauer et al. studied gramillin, secreted by the fungal pathogen Fusarium graminearum (causal agent of fusarium head blight), whereas in Nature Communications, Getzke and Wang et al. reported brassicapeptin A from the opportunistic bacterial pathogen Pseudomonas brassicacearum R401. Interestingly, both cyclic lipopeptides disrupt ionic balance in host plants through pore formation, with marginal suppression of host immunity. Specifically, gramillin induces reactive oxygen species (ROS) burst depending on host ILK1 and RBOHD genes but promotes disease independently of the major immune signaling pathways (such as jasmonic acid, salicylic acid, and pattern recognition receptors/PRRs). On the other hand, brassicapeptin A is salt-inducible and exhibits detrimental effects by interacting with salt in the environment, disturbing plant ion homeostasis. Together, these studies establish the multifaceted role of cyclic lipopeptides and their interactions with environmental stressors in pathogen success. (Summary by Arijit Mukherjee @ArijitM61745830) Cell Reports 10.1016/j.celrep.2024.114384, Nature Comms 10.1038/s41467-024-48517-5
Receptor-like cytoplasmic kinases of different subfamilies differentially regulate immune responses
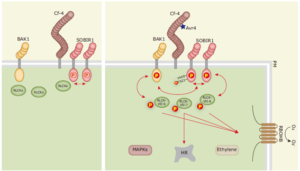 Cell surface receptor complexes act as the first line of defense in detecting pathogens and preventing invasion. Upon recognizing extracellular immunogenic patterns, a cascade of signaling relays occurs, mediated by phosphorylation events among a large array of membrane-associated proteins. These proteins include receptor-like kinases (RLKs), receptor-like proteins (RLPs), and receptor-like cytoplasmic kinases (RLCKs). The genomes of Arabidopsis and rice contain approximately 610 and 1,100 RLKs, respectively, though the functions of many of these proteins remain largely unknown. Despite the complex nature and origins of the patterns perceived, immune responses generally converge on a few common pathways. These include a rapid burst of reactive oxygen species (ROS), activation of mitogen-activated protein kinase (MAPK) cascades, transcriptional reprogramming, and, in some cases, programmed cell death known as the hypersensitive response (HR). Focusing on the RLCK subfamily VII, Huang and colleagues demonstrated that RLCK-VII-6, -7, and -8 are non-redundantly involved in the ROS burst upon detecting diverse patterns from both bacterial and fungal origins. Interestingly, only RLCK-VII-7 is involved in the HR response, highlighting the specific functions of RLCK subfamily members. (Summary by Ching Chan @ntnuchanlab) Nature Communications 10.1038/s41467-024-48313-1
Cell surface receptor complexes act as the first line of defense in detecting pathogens and preventing invasion. Upon recognizing extracellular immunogenic patterns, a cascade of signaling relays occurs, mediated by phosphorylation events among a large array of membrane-associated proteins. These proteins include receptor-like kinases (RLKs), receptor-like proteins (RLPs), and receptor-like cytoplasmic kinases (RLCKs). The genomes of Arabidopsis and rice contain approximately 610 and 1,100 RLKs, respectively, though the functions of many of these proteins remain largely unknown. Despite the complex nature and origins of the patterns perceived, immune responses generally converge on a few common pathways. These include a rapid burst of reactive oxygen species (ROS), activation of mitogen-activated protein kinase (MAPK) cascades, transcriptional reprogramming, and, in some cases, programmed cell death known as the hypersensitive response (HR). Focusing on the RLCK subfamily VII, Huang and colleagues demonstrated that RLCK-VII-6, -7, and -8 are non-redundantly involved in the ROS burst upon detecting diverse patterns from both bacterial and fungal origins. Interestingly, only RLCK-VII-7 is involved in the HR response, highlighting the specific functions of RLCK subfamily members. (Summary by Ching Chan @ntnuchanlab) Nature Communications 10.1038/s41467-024-48313-1
Alternative splicing of a disease resistance gene maintains homeostasis between growth and immunity
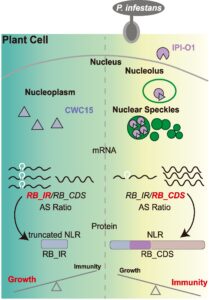 Plant resistance genes encode proteins that trigger immune responses when they recognize pathogen effectors. Their activation must be carefully regulated, as overexpression of activation of R genes usually causes a decrease in growth rate. Here, Sun et al. investigated the role of alternative splicing of a potato R gene called RB. They showed that in the absence of the Phythophtora infestans effector IPI-O1, the RB gene is spliced in such a way that an intron is retained, leading to a truncated, non-functional protein. However in the presence of the pathogen effector, the transcript is spliced to remove the intron, leading to a functional resistance protein. The authors showed that the IPI-O1 pathogen effector interacts with the plant splicing factor CWC15, causing it to localize to nuclear speckles and leading to increased splicing of the intron, functional protein production, and enhanced immunity. This finding sheds new light on how plants balance the trade-off between defense and growth. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae189
Plant resistance genes encode proteins that trigger immune responses when they recognize pathogen effectors. Their activation must be carefully regulated, as overexpression of activation of R genes usually causes a decrease in growth rate. Here, Sun et al. investigated the role of alternative splicing of a potato R gene called RB. They showed that in the absence of the Phythophtora infestans effector IPI-O1, the RB gene is spliced in such a way that an intron is retained, leading to a truncated, non-functional protein. However in the presence of the pathogen effector, the transcript is spliced to remove the intron, leading to a functional resistance protein. The authors showed that the IPI-O1 pathogen effector interacts with the plant splicing factor CWC15, causing it to localize to nuclear speckles and leading to increased splicing of the intron, functional protein production, and enhanced immunity. This finding sheds new light on how plants balance the trade-off between defense and growth. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae189