Plant Science Research Weekly: July 24
Guest Editor:

Humberto is a postdoctoral Fellow at the de Folter Lab, in CINVESTAV-Irapuato (Mexico). He holds a PhD in Plant Biotechnology. During his career, he has focussed on the study of transcription factors guiding gynoecium and fruit development in Arabidopsis. His current work is aimed in the understanding of the physical interaction dynamics of transcription factors during reproductive development.
Twitter: @herrera_h
REVIEW: Shaping Organs: Shared Structural Principles Across Kingdoms
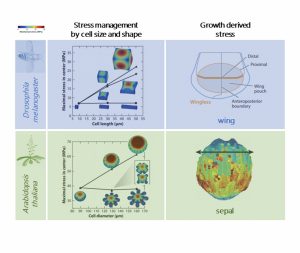
Understanding how a single fertilized cell develops into a complex multicellular system has always been a challenging but fascinating topic of developmental biology. In both animal and plant species, it all starts with a simple spherical cell; afterward, two axes of polarity generate flat shapes, and a third axis originates folded shapes. The combination of these basic structures with tissue deformations (and growth) accounts for the development of multiple functional organs in adult organisms. In this article, Hamant and Saunders reviewed recent findings on biochemical and biophysical mechanisms governing morphogenesis in model organisms and inferred general principles underpinning organ formation across kingdoms. The authors revised fundamental cellular processes – such as cell diffusion, cell adhesion, and cell polarity- directing tissue deformation, as well as biomechanical processes monitoring organ geometry. They emphasized that animal and plant systems share similar regulatory mechanisms to shape organs, although they differ in cellular structures and molecular components. A clear example comes from mechanical feedback mechanisms underlying the development of wing disc in Drosophila melanogaster and sepal in Arabidopsis thaliana. In these model systems, mechanical stress inhibits cell proliferation at the boundary with neighboring cells or tissues, ultimately determining the final size and shape of these organs. To conclude, the authors commented on many biological questions still unanswered and how new technological advances and multidisciplinary approaches can benefit research in this field. (Summary and image adaptation by Michela Osnato (@michela_osnato) Annu. Rev. Cell Dev. Biol 10.1146/annurev-cellbio-012820-103850
Cold, but not freezing: the ICE-L genome
When sea water freezes, it forms not a solid mass of ice, but rather a frozen matrix criss-crossed by tiny brine-filled channels. It is within these brine channels in the Antarctic sea ice that the psychrophilic (cold-loving) green alga ICE-L lives. How is it able to thrive in such a seemingly hostile environment of extreme cold, high salinity and large swings in light intensity and quality? The new genome assembly published by Zhang et al. offers some insights. The authors compared the ICE-L genome to other algae within the Chlamydomonadales, including both psychrophilic and mesophilic species, and found an expansion of several gene families that may play a role in adaptation to a cold and saline environment. The expanded families include, among others, fatty acid desaturases that help maintain membrane fluidity at freezing temperatures, and enzymes that help regulate ion homeostasis. They also discovered ice-binding proteins (secreted proteins that prevent freezing damage) of bacterial origin, demonstrating the importance of horizontal gene transfer in the evolution of ICE-L. The rich genetic and genomic resources available in the Chlamydomonadales are well suited for continued studies on the molecular mechanisms and evolution of cold tolerance in green algae. (Summary by Frej Tulin @FrejTulin) Curr. Biol. 10.1016/j.cub.2020.06.029
Epicotyl morphophysiological dormancy and storage behaviour of seeds of Strychnos species
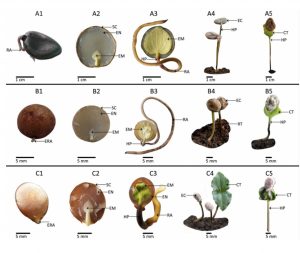
Seed dormancy and desiccation tolerance impact on germination timing and soil seed bank formation. Here, Muthuthanthirige et al. conducted a thorough series of experiments and anatomical observations to determine the dormancy class and desiccation tolerance of three Strychnos (Loganiacae) species: S. nux-vomica, S. potatorum, and S. benthamii. (As indicated by the name, plants in this genus produce toxic alkaoloids including strychnine and curare, the poison arrow toxin). All seeds had underdeveloped embryos at dispersal time. Moreover, seeds exhibited a delay in germination or shoot emergence (t50> 30 days), an aspect that was improved with gibberellin application. Consequently, seeds were classified as epicotyl morphophysiologically dormant (eMPD), being the first record of this dormancy class in the Loganiaceae. S. nux-vomica and S. potamorum seeds remained viable after desiccation, while those of S. benthamii completely lost viability. These results partially matched the authors’ hypothesis, considering S. nux-vomica and S. potamorum inhabit seasonally dry forests, where dormancy restricts germination to the rainy season, and desiccation tolerance allows seeds to survive the dry season. However, for the rainforest species, S. benthamii, epicotyl dormancy was unexpected. The authors highlight that eMPD allows seedling emergence under the most suitable conditions, which might not necessarily be the same for germination. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000203
REVIEW: Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding
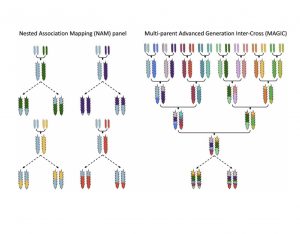
Biparental populations (BPPs) obtained by crossing two diverse inbred lines have long been used for dissecting the complex traits owing to their simplicity, ease of development and high power of detecting QTLs with a few hundred markers genotyped. Nonetheless, poor resolution and low genetic diversity of BPPs affect the precision of QTL mapping. This review by Scott et al. is a comprehensive narration of how the second generation mapping populations, the multi-parent populations (MPPs) developed by inter-crossing multiple parents, address the limitations of BPPs. The expanded genetic diversity coupled with minimal population structure due to inter-mating among multiple founders provide high resolution to QTL mapping experiments, which has promoted the popularity and uptake of MPPs; presently MPPs are available for almost all the major crops. Careful choice of founders is a key in determining the extent of genetic variation available in the population. While MPPs involving elite varieties as founders directly offer pre-breeding lines or superior breeding lines for varietal development, MPPs with landraces and wild relatives are rich source of novel allelic combinations and are valuable sources for dissecting complex traits. Next generation sequencing technologies can be adopted for whole genome re-sequencing, and de novo assembly of the founder lines would help in gene cataloguing and re-annotating the gene models. High density genotyping with novel and existing markers through high-throughput genotyping technologies would escalate further the power and precision of QTL mapping and increase the chance of landing on the causal gene. Availability of high-throughput phenotyping platforms enables multi-trait analyses, trait – trait interactions and multi-environment analyses of MPPs. Overall, MPPs with extensive genotypic and phenotypic data are valuable genetic resources that can be shared across the labs for genetic analyses of complex traits. (Summary by Haritha Bollinedi @HarithaBolline1) Heredity 10.1038/s41437-020-0336-6
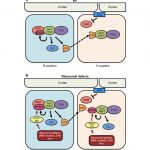
Cell fate switch in response to impaired ribosome biogenesis
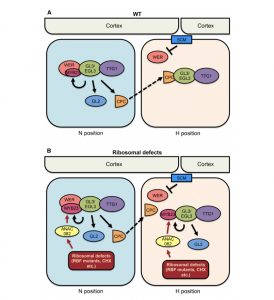
Root hairs are epidermal projections that help plants to absorb water and nutrients from the soil. Root hair development is initiated by the cumulative action of TRANSPARENT TESTA GLABRA1 (TTG1, a WD40 domain protein), GLABRA3/ENHANCER OF GLABRA3 (GL3/EGL3, a bHLH), CAPRICE (CPC, a MYB), inhibited by the same set of proteins except that CPC is replaced by other MYB protein(s) called WEREWOLF (WER) and MYB23. In the cpc mutant, the root hair production is reduced because of more ectopic non-hair cell formation, but not abolished completely, raising a question of redundancy. In this paper, Wang et al. identified a redundant root hair development pathway initiated in response to the impaired ribosome biogenesis. In an enhancer screen of the cpc mutant, the authors identified ARABIDOPSIS PUMILIO23 (APUM23), which encodes a ribosome biogenesis factor (RBF) and positively regulates root hair development. Using molecular genetic experiments, the authors found that apum23 shows an ectopic expression of MYB23, mediated by ANAC082 (encoding a NAC family transcription factor). Also, the authors identified ectopic non-hair cell formation in other RBF mutants and in response to flaellin22, suggesting that epidermal cell fate switch is a general response to RBF deficiency and few biotic stresses. Thus, this paper advances our understanding of cell fate switch between root hair and non-hair cells in responses to various stress conditions by activating ANAC082 and MYB23 pathway. (Summary by Vijaya Batthula @Vijaya_Batthula) The Plant Cell 10.1105/tpc.19.00773
Temperature-dependent growth contributes to long-term cold sensing
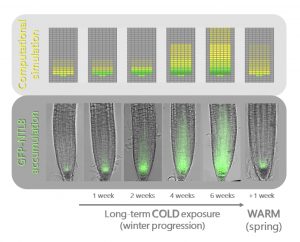
Seasonal changes in ambient temperature greatly influence plant development. While the response to acute heat stress has been widely studied, the regulatory mechanisms that integrate naturally fluctuating temperatures – such as the progression of winter – remain largely unknown. This paper sheds new light on the activation of VERNALIZATION INSENSITIVE 3 (VIN3), a key regulator of the response to long-term cold exposure in Arabidopsis. Zhao and colleagues mutagenized a reporter line carrying VIN3 fused to LUCIFERASE (in vin3-6 background) and identified dominant mutations in NTL8, encoding a membrane-associated NAC transcription factor, with altered VIN3-LUC expression patterns. The analysis of multiple GFP–NTL8 reporter lines indicated that the fluorescence signal increased gradually in root meristems in response to cold conditions and decreased in post-cold warm conditions. Rather than changes in transcript levels, low protein turnover contributed to NTL8 accumulation during vernalization, and consequently a progressive increase of VIN3 expression. Modeling combined with experimental validation demonstrated that growth-dependent protein dilution determined NTL8 accumulation: faster growth at warm temperatures associated with lower NTL8 amounts, whereas slower growth at low temperatures with higher NTL8 amounts. Similar patterns were also observed in plants subjected to non -cold treatments affecting plant development: lower NTL8 concentrations upon treatment with growth-promoting hormones, and higher NTL concentrations with growth inhibiting compounds. To conclude, seasonal changes can be sensed through indirect mechanisms relying on protein dilution mediated by temperature-dependent growth. This long-term temperature-sensing system can be applied to other long-lived proteins with a constant production and slow degradation. (Summary and image adaptation by Michela Osnato @michela_osnato) Nature 10.1038/s41586-020-2485-4
Rubisco accumulation factor 1 and carboxysome biogenesis
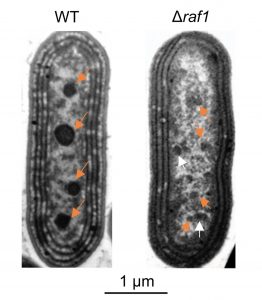
Carbon concentration mechanisms (CCMs) refer to a diverse set of strategies by which photosynthetic organisms increase the amount of carbon dioxide available to the carbon-fixing enzyme Rubisco. The cyanobacterial CCM relies on the carboxysome, a membraneless microcompartment with a core of densely packed Rubisco holoenzymes surrounded by a proteinaceous shell. Each holoenzyme is composed of four anti-parallel Rubisco large subunit (RbcL) dimers together with eight small subunits (RbcS). The Rubisco Assembly Factor 1 (Raf1) is known to assist in Rubisco holoenzyme assembly during the early stages of carboxysome formation. Here, Huang et al. use cryo-electron microscopy to solve the structure of the RbcL-Raf1 complex, showing how Raf1 assists in both RbcL dimer formation and in dimer-dimer interactions, thereby stabilizing the holoenzyme. This job is apparently critical, because in the absence of Raf1 carboxysome formation is severely disrupted and the cells grow poorly unless supplemented with high concentration of carbon dioxide. (Summary by Frej Tulin @FrejTulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2007990117
A new miRNA complex assembly partner – Intrinsically disordered but an important scaffold
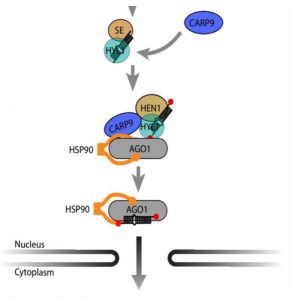
Gene regulation through microRNAs (miRNA) starts in the nucleus with transcription and processing into mature miRNA duplexes. Loading of miRNA into ARGONAUTE1 (AGO1) and the assembly of the RNA-Induced Silencing Complex was thought to be exclusively a cytoplasmic process, but new research reports this also happens in the nucleus through unknown mechanisms. Tomassi, Re, and colleagues identified CONSTITUTIVE ALTERATIONS IN THE SMALL RNAS PATHWAYS9 (CARP9) as a new partner in the miRNA pathway. CARP9 is an intrinsically disordered protein that localizes to the nucleus; CARP9 paralogs are also predicted to be disordered in all plant species. Mutations in CARP9 produced morphological alterations, a mild reduction in the miRNA accumulation, and impaired gene silencing. The authors discovered that CARP9 interacts with HYPONASTIC LEAVES1 (HYL1) to promote HYL1-AGO1 interactions for loading mature miRNA into AGO1. The data suggest that CARP9 acts as a scaffold for the formation of a nuclear post pri-miRNA processing complex to allow proper loading of miRNA and, regulation of gene expression. (Summary by Katy Dunning @plantmomkaty) Plant Physiology 10.1104/pp.20.00258
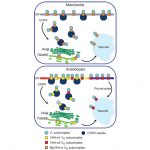
Trafficking of VHA-a isoforms in Arabidopsis thaliana

Vacuolar-type ATPase (V-ATPase) is a highly conserved enzyme but functionally diverse among eukaryotic cells. V-ATPases couple the energy released by ATP hydrolysis to pump protons across membranes of many cell types. In Arabidopsis, the V-ATPases containing different isoforms of V-ATPase catalytic subunit A (VHA-a) are sorted in the endoplasmic reticulum (ER); however, it is not clear how these isoforms are targeted in the cell. Lupanga et al. identified a VHA-a1 targeting domain (a1-TD) that is conserved among seed plants; this a1-TD is required for COPII-mediated ER-export and retention in trans-Golgi network/early endosome (TGN/EE). When mutated in the VHA-a1-TD residue, this VHA-a1 subunit could partially replace the tonoplast V-ATPase (VHA-a2 and VHA-a3). By contrast, the liverwort Marchantia (a non-seed plant), has only single copy of VHA-a, which is predominantly localized to the tonoplast and partially co-localized at the TGN/EE. Interestingly, the predicted-to-be lethal null alleles of VHA-a1 have a dwarf phenotype. Further studies showed the function of VHA-a1 is critical for pollen development but can be replaced by VHA-a2 and VHA-a3 during vegetative growth. Overall, this study found a novel targeting motif for V-ATPase and revealed its essential function for pollen development. (Summary by Min May Wong @wongminmay) bioRxiv 10.1101/2020.06.30.179333
Ancient seeds reconstruction and the evolution of integuments
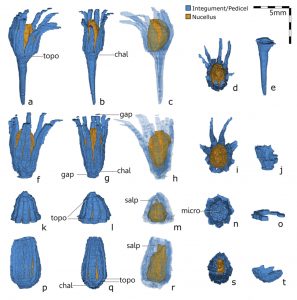
The origin of seeds – the specialized structures that contain and protect the developing embryos- is a key event in plant evolution. Primitive seeds comprise an exposed nucellus surrounded by a lobate integument; in contrast, extant seeds have one or more integuments fully enclosing the nucellus. To understand the evolution of integuments, Meade and colleagues analyzed Genomosperma fossils; seeds in this species represent the most primitive stages of integument organization. Authors re-examined samples of two Genomosperma species, G. kidstoni and G. latens. They used serial sections of samples to reconstruct high-quality 3D models of seeds of the two species. Reconstructions of Genomosperma allowed them to examine morphology with high confidence and accuracy, resolving the structure and organization of integument, pedicel, and nucellus in Genomosperma seeds. These models confirmed some previous 2D morphological studies; additional morphological features were measured and quantitatively analyzed within the two species samples. The authors found no anatomical differences between G. kidstoniand G. latens, so conclude both belong to a single species: G kidstoni. On the other hand, significant differences were found in the organization, flexure, and fusion of the integumentary lobes. The observation of variations in the integument lobe number suggests that the formation of Genomosperma integuments could be similar to the formation of organs within a flower; in this case, there could also be a variation of organ number when the expression of boundary genes is altered. The authors are currently analyzing the biomechanical properties of integumentary lobes to better understand the function of Genomosperma integuments. (Summary by Humberto Herrera-Ubaldo) New Phytologist 10.1111/nph.16792