Plant Science Research Weekly: July 19th
Review: Formal description of plant morphogenesis ($)
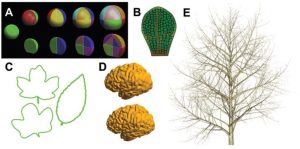 In recent years, a number of tools have been developed to describe and model plant morphogenesis, and these approaches have greatly informed our understanding of the molecular processes that underpin the control of growth. This excellent review by Pałubicki et al. is “an attempt to bring together a large number of computational modeling concepts and make them accessible to the analytical as well as empirical student of plant morphogenesis,” and it works. The authors point out that growth is the result of signals that specify changes over time, and that describing growth depends on having tools to represent both shape and information flow. They summarize (with pictures) different methods for describing plant geometry and signaling, and present the strength and limitations of each approach. I found this review particularly helpful in my efforts to understand the ideas and terms used in this developing field. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz210
In recent years, a number of tools have been developed to describe and model plant morphogenesis, and these approaches have greatly informed our understanding of the molecular processes that underpin the control of growth. This excellent review by Pałubicki et al. is “an attempt to bring together a large number of computational modeling concepts and make them accessible to the analytical as well as empirical student of plant morphogenesis,” and it works. The authors point out that growth is the result of signals that specify changes over time, and that describing growth depends on having tools to represent both shape and information flow. They summarize (with pictures) different methods for describing plant geometry and signaling, and present the strength and limitations of each approach. I found this review particularly helpful in my efforts to understand the ideas and terms used in this developing field. (Summary by Mary Williams) J. Exp. Bot. 10.1093/jxb/erz210
GMO-free RNAi: exogenous application of RNA molecules in plants
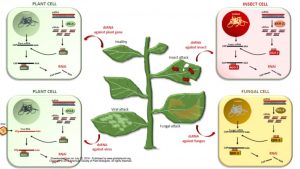 Criticism of transgenic plants and GMOs motivates research into effective GMO-free RNA delivery methods. In this review, Dalakouras et al. discuss different strategies for exogenous application of RNA molecules (dsRNAs, siRNAs) into plants to trigger RNA interference (RNAi) against various targets, such as endogenous plant genes, viruses, viroids, insects, mites and fungi. The authors conclude that the most efficient way to induce systemic RNAi in plants is by exogenously applying 22-nt siRNAs. For plant gene and virus targets, high-pressure spraying allows symplastic RNA delivery, enabling RNAi uptake into plant cells for endogenous gene silencing. For insect and fungus management, because RNAi takes place inside their cells, the uptake of non-processed dsRNA is needed. Trunk injection, petiole absorption and low-pressure spraying as apoplastic delivery methods allow this. Furthermore, secretion of exosome-like extracellular vesicles in Arabidopsis has been identified, which enables RNA delivery into other taxonomic kingdom such as fungal cells. Finally, the authors considered technical improvements for dsRNA applications, like joining them with nanoparticles and carrier peptides to improve delivery, and using large-scale bacterial bioreactors to lower the costs of dsRNA production. (Summary by Ana Valladares). Plant Physiol. 10.1104/pp.19.00570
Criticism of transgenic plants and GMOs motivates research into effective GMO-free RNA delivery methods. In this review, Dalakouras et al. discuss different strategies for exogenous application of RNA molecules (dsRNAs, siRNAs) into plants to trigger RNA interference (RNAi) against various targets, such as endogenous plant genes, viruses, viroids, insects, mites and fungi. The authors conclude that the most efficient way to induce systemic RNAi in plants is by exogenously applying 22-nt siRNAs. For plant gene and virus targets, high-pressure spraying allows symplastic RNA delivery, enabling RNAi uptake into plant cells for endogenous gene silencing. For insect and fungus management, because RNAi takes place inside their cells, the uptake of non-processed dsRNA is needed. Trunk injection, petiole absorption and low-pressure spraying as apoplastic delivery methods allow this. Furthermore, secretion of exosome-like extracellular vesicles in Arabidopsis has been identified, which enables RNA delivery into other taxonomic kingdom such as fungal cells. Finally, the authors considered technical improvements for dsRNA applications, like joining them with nanoparticles and carrier peptides to improve delivery, and using large-scale bacterial bioreactors to lower the costs of dsRNA production. (Summary by Ana Valladares). Plant Physiol. 10.1104/pp.19.00570
Cryo-EM structure of OSCA1.2 sheds light on the mechanical basis of membrane hyperosmolality gating ($)
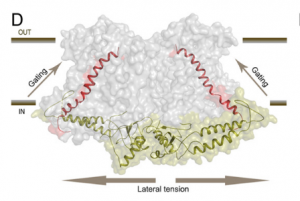 Osmotic stress in plants elicits many responses, one of which is increased accumulation of Ca2+ in the cytosol. Genes involved in this response have been identified, yet the mechanism behind the Ca2+ transport remains unknown. Maity et al. investigated the structure and function of the osmolality-sensitive channel OSCA1.2, a candidate for hyperosmolarity-induced Ca2+ transport, using cryo-electron microscopy along with reconstitution of the protein in droplet-interfaced bilayers. They demonstrated that OSCA1.2 is dimeric, with each monomer containing 11 transmembrane domains (TMDs), as well as a large cytosolic loop between TM2 and TM3 that mediates the dimeric interaction. The predicted transport pathway contains 5 amino acids contributed from TM5 and TM6. Based on computer simulations, these may interact with the cytosolic loop to sense conformational changes that can be translated to transport activity under hyperosmotic scenarios. However, a Ca2+ fluorescent dye and two-electrode voltage clamping with Xenopus oocytes didn’t provide evidence of osmotic stress-induced Ca2+ flux. Therefore, substrate identity and the gating mechanism of OSCA1.2 remain questions for future research. (Summary by Nathan Scinto-Madonich) PNAS 10.1073/pnas.1900774116 .
Osmotic stress in plants elicits many responses, one of which is increased accumulation of Ca2+ in the cytosol. Genes involved in this response have been identified, yet the mechanism behind the Ca2+ transport remains unknown. Maity et al. investigated the structure and function of the osmolality-sensitive channel OSCA1.2, a candidate for hyperosmolarity-induced Ca2+ transport, using cryo-electron microscopy along with reconstitution of the protein in droplet-interfaced bilayers. They demonstrated that OSCA1.2 is dimeric, with each monomer containing 11 transmembrane domains (TMDs), as well as a large cytosolic loop between TM2 and TM3 that mediates the dimeric interaction. The predicted transport pathway contains 5 amino acids contributed from TM5 and TM6. Based on computer simulations, these may interact with the cytosolic loop to sense conformational changes that can be translated to transport activity under hyperosmotic scenarios. However, a Ca2+ fluorescent dye and two-electrode voltage clamping with Xenopus oocytes didn’t provide evidence of osmotic stress-induced Ca2+ flux. Therefore, substrate identity and the gating mechanism of OSCA1.2 remain questions for future research. (Summary by Nathan Scinto-Madonich) PNAS 10.1073/pnas.1900774116 .
Mechanisms of RALF peptide perception by a heterotypic receptor complex ($)
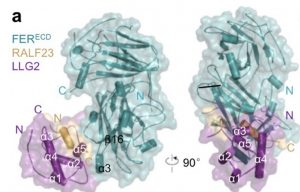 RALFs (Rapid Alkalinization Factors, based on historical observations that they induce alkalization of the extracellular space) are signaling peptides with diverse roles. RALFs have previously been shown to bind to the extracellular domain of (among others) FERONIA (FER), a member of the Catharanthus roseus RLK1-like (CrRLK1L) family. LORELIE-Like GPI-membrane anchored protein (LLG) has been shown to operate in the FER signaling pathway. Now, Xiao et al. have found that RALF23 binds to a complex formed by FER and LLG. They also identified an N-terminal “YISY” sequence that is conserved in other RALFs and that is important for this interaction. Their data also suggest that the RALF+FER can interact with other LLGs, which could explain the diverse processes that RALFs and FER are involved in. (Summary by Mary Williams) Nature 10.1038/s41586-019-1409-7
RALFs (Rapid Alkalinization Factors, based on historical observations that they induce alkalization of the extracellular space) are signaling peptides with diverse roles. RALFs have previously been shown to bind to the extracellular domain of (among others) FERONIA (FER), a member of the Catharanthus roseus RLK1-like (CrRLK1L) family. LORELIE-Like GPI-membrane anchored protein (LLG) has been shown to operate in the FER signaling pathway. Now, Xiao et al. have found that RALF23 binds to a complex formed by FER and LLG. They also identified an N-terminal “YISY” sequence that is conserved in other RALFs and that is important for this interaction. Their data also suggest that the RALF+FER can interact with other LLGs, which could explain the diverse processes that RALFs and FER are involved in. (Summary by Mary Williams) Nature 10.1038/s41586-019-1409-7
Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth
 RALFs (Rapid Alkalinization Factors, based on historical observations that they induce alkalization of the extracellular space) are signaling peptides with diverse roles. RALF4 is required for pollen tube growth and guidance through its interactions with CrRLK1Ls and leucine-rich extension (LRX) proteins. Moussu, Broyart et al. characterized the complex formed between RALF4 and homodimers of the LRX2 or LRX8. RALF4 has conserved Cys residues and binds to the LRR core of the LRXs in the folded conformation. The RALF binding pocket is highly conserved among the known LRX family members. The authors also examined binding of RALF4 with pollen-expressed CrRLK1Ls and found that these interactions occur with very different affinities and modes of binding. The authors conclude, “ these findings suggest that RALFs have the capacity to instruct different signaling proteins to coordinate cell wall remodeling during pollen tube growth.” (Summary by Mary Williams) bioRxiv 10.1101/695874
RALFs (Rapid Alkalinization Factors, based on historical observations that they induce alkalization of the extracellular space) are signaling peptides with diverse roles. RALF4 is required for pollen tube growth and guidance through its interactions with CrRLK1Ls and leucine-rich extension (LRX) proteins. Moussu, Broyart et al. characterized the complex formed between RALF4 and homodimers of the LRX2 or LRX8. RALF4 has conserved Cys residues and binds to the LRR core of the LRXs in the folded conformation. The RALF binding pocket is highly conserved among the known LRX family members. The authors also examined binding of RALF4 with pollen-expressed CrRLK1Ls and found that these interactions occur with very different affinities and modes of binding. The authors conclude, “ these findings suggest that RALFs have the capacity to instruct different signaling proteins to coordinate cell wall remodeling during pollen tube growth.” (Summary by Mary Williams) bioRxiv 10.1101/695874
Recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2
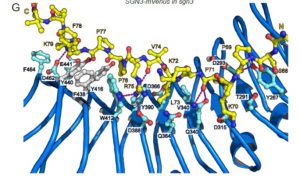 SCHENGEN 3 [SGN3, also known as GASSHO1 (GSO1)] is a leucine-rich repeat receptor kinase (LRR-RK) previously shown to interact with CIF peptides (CASPARIAN STRIP INTEGRITY FACTORS) to regulate the development of the Casparian strip boundary in roots. Here, Okuda et al. characterize the CIF/LRR-RK interaction. They show that that there is a wide range of binding affinities of LRR-RK / peptide ligand interactions and that GSO1/SGN3 forms the strongest receptor-ligand interactions identified in plantshas and has a longer LRR domain and peptide binding domain than other LRR-RKs. Mutational studies indicate that this strong interaction is mediated by a large number of residues., and that multiple mutations are required to disrupt it. Additionally, the authors identified two previously unannotated CIF proteins and implicate SERK LRR-RKs as essential co-receptor kinases. (Summary by Mary Williams) bioRxiv 10.1101/692228
SCHENGEN 3 [SGN3, also known as GASSHO1 (GSO1)] is a leucine-rich repeat receptor kinase (LRR-RK) previously shown to interact with CIF peptides (CASPARIAN STRIP INTEGRITY FACTORS) to regulate the development of the Casparian strip boundary in roots. Here, Okuda et al. characterize the CIF/LRR-RK interaction. They show that that there is a wide range of binding affinities of LRR-RK / peptide ligand interactions and that GSO1/SGN3 forms the strongest receptor-ligand interactions identified in plantshas and has a longer LRR domain and peptide binding domain than other LRR-RKs. Mutational studies indicate that this strong interaction is mediated by a large number of residues., and that multiple mutations are required to disrupt it. Additionally, the authors identified two previously unannotated CIF proteins and implicate SERK LRR-RKs as essential co-receptor kinases. (Summary by Mary Williams) bioRxiv 10.1101/692228
Root system depth is shaped by EXOCYST70A3 via modulation of auxin transport
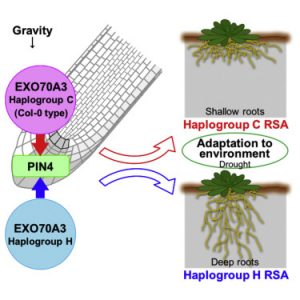
The angle at which roots grow into the soil is modulated by the interaction between genetics and environment, and involves the gravity perception and response pathways including auxin transporters. Ogura et al. did a GWAS analysis of the gravitropic response of different Arabidopsis accessions in the presence of the auxin-transport inhibitor NPA, and identified allelic variation in EXOCYST70A3, encoding an exocytosis factor, as involved in root response to NPA. Specifically, perturbation of EXO70A3 expression regulates PIN4 levels and patterns in root columella cells, hence auxin distribution. In plants grown in pots, allelic variation or altered expression of EXO70A3 affected root system architecture. Furthermore, one of the naturally occurring haplotypes contributed to higher seed set under drought conditions. As part of a larger effort, this gene can contribute to efforts to engineer deep, carbon-storing roots to slow the rate of anthropomorphic climate change. (Summary by Mary Williams) Cell 10.1016/j.cell.2019.06.021
Conserved biochemical defenses underpin host responses to oomycete infection in liverwort
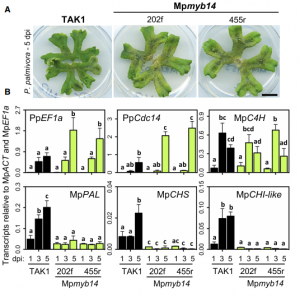 Marchantia polymorpha is an emerging model for plant molecular biology and has contributed to studies on development and plant-microbe interactions. Here, using RNA-seq and proteomics, Carella et al. present a detailed time-course analysis of Marchantia molecular responses to pathogen infection triggered by the oomycete Phytophtora palmitovora. By comparing Marchantia to tobacco responses, the authors show that many features of the molecular defense responses are conserved across land plants. Among them, genes encoding Pathogenesis-related (PR) proteins, phenylpropanoid biosynthesis enzymes, and transcription factors are induced after infection. As part of the response, anthocyanin-like compounds are synthesized and accumulated around Marchantia air chambers during infection. Altering expression of MpMyb14, a transcription factor that it is induced by pathogenesis and regulates flavonoid biosynthesis, alters susceptibity or resistance to P. palmitovora, demonstrating that flavonoid biosynthesis is an ancient and conserved defense mechanism and works in Marchantia similarly to angiosperms. This paper, along with that of Gimenez-Ibanez et al., establish Marchantia-microbe interaction systems that will serve as powerful tools to dissect plant defense mechanisms. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.05.078.
Marchantia polymorpha is an emerging model for plant molecular biology and has contributed to studies on development and plant-microbe interactions. Here, using RNA-seq and proteomics, Carella et al. present a detailed time-course analysis of Marchantia molecular responses to pathogen infection triggered by the oomycete Phytophtora palmitovora. By comparing Marchantia to tobacco responses, the authors show that many features of the molecular defense responses are conserved across land plants. Among them, genes encoding Pathogenesis-related (PR) proteins, phenylpropanoid biosynthesis enzymes, and transcription factors are induced after infection. As part of the response, anthocyanin-like compounds are synthesized and accumulated around Marchantia air chambers during infection. Altering expression of MpMyb14, a transcription factor that it is induced by pathogenesis and regulates flavonoid biosynthesis, alters susceptibity or resistance to P. palmitovora, demonstrating that flavonoid biosynthesis is an ancient and conserved defense mechanism and works in Marchantia similarly to angiosperms. This paper, along with that of Gimenez-Ibanez et al., establish Marchantia-microbe interaction systems that will serve as powerful tools to dissect plant defense mechanisms. (Summary by Facundo Romani) Curr. Biol. 10.1016/j.cub.2019.05.078.
A liverwort-Pseudomonas interaction reveals an ancient plant defensive mechanism
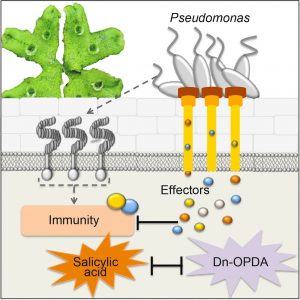 Plants are sessile organisms that have evolved sophisticated immune systems in defense of pathogens, thus maximizing the chance of survival. Most of our understanding of plant defenses comes from studies in angiosperms. Evolutionary molecular plant-microbe interactions (EvoMPMI) can reveal the origins of plant defenses, but have been limited by a lack of suitable pathosystems in in basal land plants. Recently Gimenez-Ibanez et al. uncovered an interaction between the bacterial pathogen P. syringae pv tomato (Pto) DC3000 and the liverwort Marchantia polumorpha. When exposed to Pto DC3000, M. polumorpha activates typical hallmarks of pattern-triggered immunity. Bacterial effectors such as AvrPtoA or AvrPtoB are necessary to for virulence, and the interaction depends on the genetic background of host and pathogen. This work shows that the microbe-plant interaction is ancient and highly conserved from early land plant to angiosperms, and sheds lights on novel defensive mechanism hidden by gene redundancy in later-diverged plants. In summary, these discoveries, along with that of Carella et al., provide novel experimental systems to study EvoMPMI, and help us understanding the co-evolution of plants and pathogens during land plant colonization. (Summarized by Nanxun Qin) Curr. Biol. 10.1016/j.cub.2019.05.079
Plants are sessile organisms that have evolved sophisticated immune systems in defense of pathogens, thus maximizing the chance of survival. Most of our understanding of plant defenses comes from studies in angiosperms. Evolutionary molecular plant-microbe interactions (EvoMPMI) can reveal the origins of plant defenses, but have been limited by a lack of suitable pathosystems in in basal land plants. Recently Gimenez-Ibanez et al. uncovered an interaction between the bacterial pathogen P. syringae pv tomato (Pto) DC3000 and the liverwort Marchantia polumorpha. When exposed to Pto DC3000, M. polumorpha activates typical hallmarks of pattern-triggered immunity. Bacterial effectors such as AvrPtoA or AvrPtoB are necessary to for virulence, and the interaction depends on the genetic background of host and pathogen. This work shows that the microbe-plant interaction is ancient and highly conserved from early land plant to angiosperms, and sheds lights on novel defensive mechanism hidden by gene redundancy in later-diverged plants. In summary, these discoveries, along with that of Carella et al., provide novel experimental systems to study EvoMPMI, and help us understanding the co-evolution of plants and pathogens during land plant colonization. (Summarized by Nanxun Qin) Curr. Biol. 10.1016/j.cub.2019.05.079
Forces required for Venus flytrap trigger hairs to detect small insect prey ($)
 The remarkable adaptations of Venus flytrap traps enable them to sense and respond to insects, snapping shut to capture and then digest the unfortunate meal. Previous studies showed that the sensors, trigger hairs on the inner surface of the leaves, need at least two touches to initiate trap closure. New work by Scherzer et al. refines our understanding of this process. The authors measured the force that must be exerted on the trigger hairs, in terms of how much and how quickly they must be displaced, and the weight of the prey required for such movement. Their findings indicate that although most prey trigger trap closure, some tiny prey (less than 3 mg) are effectively ignored, suggesting that the payoff of such small prey might not be worth the effort to capture it. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0465-1
The remarkable adaptations of Venus flytrap traps enable them to sense and respond to insects, snapping shut to capture and then digest the unfortunate meal. Previous studies showed that the sensors, trigger hairs on the inner surface of the leaves, need at least two touches to initiate trap closure. New work by Scherzer et al. refines our understanding of this process. The authors measured the force that must be exerted on the trigger hairs, in terms of how much and how quickly they must be displaced, and the weight of the prey required for such movement. Their findings indicate that although most prey trigger trap closure, some tiny prey (less than 3 mg) are effectively ignored, suggesting that the payoff of such small prey might not be worth the effort to capture it. (Summary by Mary Williams) Nature Plants 10.1038/s41477-019-0465-1



