Plant Science Research Weekly: July 15, 2022
Review: Root cell types as an interface for biotic interactions
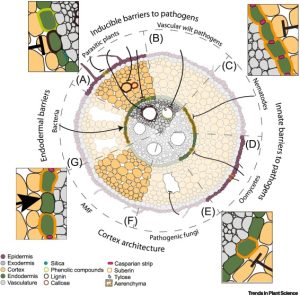 Kawa and Brady review the contributions and responses of individual cell types and cell identities in plant biotic interactions, both pathogenic and commensal. The first step in these interactions is the perception of the microbes by root cells, which involves highly conserved microbe-associated molecular patterns (MAMPs) such as flagellin. MAMPs can trigger plant defense responses, which can sometimes be suppressed or evaded by microbial processes. Transcriptional responses to MAMPs are cell-type specific (stronger in epidermal cells). Many microbes move into deeper root tissues (e.g., vasculature), but this progress can be halted or hindered by barriers including the Casparian strip. Roots can respond to pathogens by increasing the production of anti-microbial metabolites or compounds that contribute to barrier formation. Interestingly, some microbes can facilitate their entry by reprograming plant cells. Root radial anatomy is diverse in terms of numbers and types of cell layers, and these factors also affect microbial interactions. This deep and detailed dive into the very localized responses of roots to beneficial and harmful microbes raises many interesting opportunities for engineering plants to be more welcoming to helpful microbes while maintaining or strengthening their barriers to those that cause harm. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2022.06.003
Kawa and Brady review the contributions and responses of individual cell types and cell identities in plant biotic interactions, both pathogenic and commensal. The first step in these interactions is the perception of the microbes by root cells, which involves highly conserved microbe-associated molecular patterns (MAMPs) such as flagellin. MAMPs can trigger plant defense responses, which can sometimes be suppressed or evaded by microbial processes. Transcriptional responses to MAMPs are cell-type specific (stronger in epidermal cells). Many microbes move into deeper root tissues (e.g., vasculature), but this progress can be halted or hindered by barriers including the Casparian strip. Roots can respond to pathogens by increasing the production of anti-microbial metabolites or compounds that contribute to barrier formation. Interestingly, some microbes can facilitate their entry by reprograming plant cells. Root radial anatomy is diverse in terms of numbers and types of cell layers, and these factors also affect microbial interactions. This deep and detailed dive into the very localized responses of roots to beneficial and harmful microbes raises many interesting opportunities for engineering plants to be more welcoming to helpful microbes while maintaining or strengthening their barriers to those that cause harm. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2022.06.003
CHIQUITA1 maintains the temporal transition between proliferation and differentiation in Arabidopsis thaliana
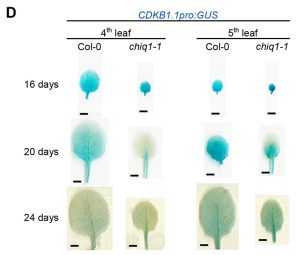 Organ size is imperative to biological function. In plants, newly-developing organs start out as a pool of meristematic cells, which set the number of cells in the organ through regulated iterations of cellular division; eventually, these cells transition out of the cell cycle and differentiate, during which they may undergo expansion/elongation. Ultimately, these two phases of development determine the parameters defining organ size: cell number and cell size. However, the molecular orchestrations through which plants control the transition from proliferation to differentiation are still mysterious. Here, Bossi et al. dissect the role of a gene previously identified as relevant to body size, CHIQUITA1 (CHIQ1), towards this transition. While fully-developed null mutants of CHIQUITA1 (chiq1-1) are dwarves, organs of chiq1-1 early in development were actually larger than those of the wild-type (WT). Leaf cell size analyses revealed this was due to younger chiq1-1 leaves having a higher proportion of differentiating, and thus expanding, cells than WT leaves of the same age. Molecular analysis of cell cycle markers show that chiq1-1 organs reduce their domains of proliferation earlier than WT organs. Measurements of cell cycle length confirm that cell cycle progression occurs normally in chiq1-1, suggesting that the cells of mutant organs exit proliferation prematurely. CHIQ1 was previously hypothesized to interact with CHIQ1-like (CHIQL) proteins. Organ and cell size analyses comparing WT, chiq1-1, and a quadruple null mutant of CHIQ1, CHIQL4, CHIQL5, and CHIQL6 indicate that CHIQ1 could be regulating proliferation exit via complexed activity with these CHIQL proteins. From this work, we attain greater insight into the genetic controls involved in plant organ development, and see the necessity of analyzing developmental phenotypes at the cellular level in addition to the organ level. (Summary by Benjamin Jin) Development 10.1242/dev.200565
Organ size is imperative to biological function. In plants, newly-developing organs start out as a pool of meristematic cells, which set the number of cells in the organ through regulated iterations of cellular division; eventually, these cells transition out of the cell cycle and differentiate, during which they may undergo expansion/elongation. Ultimately, these two phases of development determine the parameters defining organ size: cell number and cell size. However, the molecular orchestrations through which plants control the transition from proliferation to differentiation are still mysterious. Here, Bossi et al. dissect the role of a gene previously identified as relevant to body size, CHIQUITA1 (CHIQ1), towards this transition. While fully-developed null mutants of CHIQUITA1 (chiq1-1) are dwarves, organs of chiq1-1 early in development were actually larger than those of the wild-type (WT). Leaf cell size analyses revealed this was due to younger chiq1-1 leaves having a higher proportion of differentiating, and thus expanding, cells than WT leaves of the same age. Molecular analysis of cell cycle markers show that chiq1-1 organs reduce their domains of proliferation earlier than WT organs. Measurements of cell cycle length confirm that cell cycle progression occurs normally in chiq1-1, suggesting that the cells of mutant organs exit proliferation prematurely. CHIQ1 was previously hypothesized to interact with CHIQ1-like (CHIQL) proteins. Organ and cell size analyses comparing WT, chiq1-1, and a quadruple null mutant of CHIQ1, CHIQL4, CHIQL5, and CHIQL6 indicate that CHIQ1 could be regulating proliferation exit via complexed activity with these CHIQL proteins. From this work, we attain greater insight into the genetic controls involved in plant organ development, and see the necessity of analyzing developmental phenotypes at the cellular level in addition to the organ level. (Summary by Benjamin Jin) Development 10.1242/dev.200565
Leaf cell-specific and single-cell transcriptional profiling reveals a role for the palisade layer in UV light protection
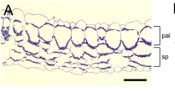 Leaves are the specialized organs of the plant that have evolved to maximize the use of light and CO2 for efficient photosynthesis. Although knowledge of photosynthesis has exploded recently, the molecular intricacies underlying leaf anatomy, especially mesophyll, remains unravelled. The palisade mesophyll forms the columnar layer at the adaxial side and conducts light into the leaf. Despite its importance in photosynthesis, less is known about the unique molecular markers of palisade cells. Procko et al. generated a transcriptional profile of palisade mesophyll in Arabidopsis through the combination of bulk palisade cell purification and single-cell RNA sequencing. Although the cells have unique cellular feature and placement, the gene expression was highly similar to other photosynthetic cells, suggesting a conserved genetic blueprint of photosynthetic cells. However, expression of genes from phenylpropanoid pathway were enriched in palisade cells. The authors revealed that a phenylpropanoid related gene FAH1 (FERULIC ACID 5-HYDROXYLASE 1) producing a UV-B protectant, sinapoylmalate, was expressed at high levels in palisade cells. This highlights the novel role of palisade mesophyll as the “sunscreen” in protecting the leaf from photooxidative UV damage that is further supported by its location. (Summary by Prakshi Aneja @PrakshiAneja) Plant Cell 10.1093/plcell/koac167
Leaves are the specialized organs of the plant that have evolved to maximize the use of light and CO2 for efficient photosynthesis. Although knowledge of photosynthesis has exploded recently, the molecular intricacies underlying leaf anatomy, especially mesophyll, remains unravelled. The palisade mesophyll forms the columnar layer at the adaxial side and conducts light into the leaf. Despite its importance in photosynthesis, less is known about the unique molecular markers of palisade cells. Procko et al. generated a transcriptional profile of palisade mesophyll in Arabidopsis through the combination of bulk palisade cell purification and single-cell RNA sequencing. Although the cells have unique cellular feature and placement, the gene expression was highly similar to other photosynthetic cells, suggesting a conserved genetic blueprint of photosynthetic cells. However, expression of genes from phenylpropanoid pathway were enriched in palisade cells. The authors revealed that a phenylpropanoid related gene FAH1 (FERULIC ACID 5-HYDROXYLASE 1) producing a UV-B protectant, sinapoylmalate, was expressed at high levels in palisade cells. This highlights the novel role of palisade mesophyll as the “sunscreen” in protecting the leaf from photooxidative UV damage that is further supported by its location. (Summary by Prakshi Aneja @PrakshiAneja) Plant Cell 10.1093/plcell/koac167
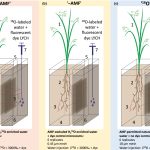
Tracking water transport to the host plants by arbuscular mycorrhizal fungi
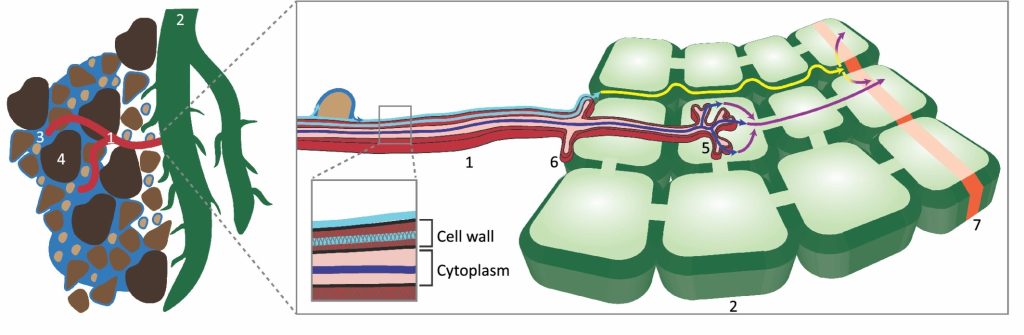 In spite of several years of observation of the symbiotic relationship between plants and arbuscular mycorrhizal fungi, we are still learning about the intricate mechanisms and processes behind their relationship. Symbiotic fungi provide nutrients (mainly phosphorus and nitrogen), stress tolerance, and water to the host plant in return for sugars and lipids. While there have been significant advances in our knowledge about the nutrient exchange mechanisms, understanding of water transportat by fungi to the host plant has remained elusive. Kakouridis et al. demonstrated that the fungal hyphae act as an extension of the root system, driving water flow along hyphae under the influence of plant transpirational pull. Using 18O-labeled water and a fluorescent dye, they demonstrated that hyphae could assist in increasing water accessibility to the roots by overcoming the discontinuity in the water film caused by air pockets or/and variable soil particle size. This study aids to our understanding of drought stress tolerance in a symbiotic system. (Summary by Chandan Kumar Gautam @chandan_gautam) New Phytol. 10.1111/nph.18281
In spite of several years of observation of the symbiotic relationship between plants and arbuscular mycorrhizal fungi, we are still learning about the intricate mechanisms and processes behind their relationship. Symbiotic fungi provide nutrients (mainly phosphorus and nitrogen), stress tolerance, and water to the host plant in return for sugars and lipids. While there have been significant advances in our knowledge about the nutrient exchange mechanisms, understanding of water transportat by fungi to the host plant has remained elusive. Kakouridis et al. demonstrated that the fungal hyphae act as an extension of the root system, driving water flow along hyphae under the influence of plant transpirational pull. Using 18O-labeled water and a fluorescent dye, they demonstrated that hyphae could assist in increasing water accessibility to the roots by overcoming the discontinuity in the water film caused by air pockets or/and variable soil particle size. This study aids to our understanding of drought stress tolerance in a symbiotic system. (Summary by Chandan Kumar Gautam @chandan_gautam) New Phytol. 10.1111/nph.18281
A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants
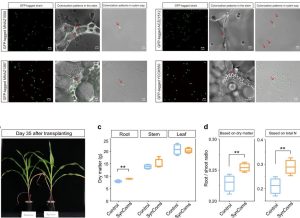 Plants and microorganisms build complex associations that are essential for plant homeostasis. Studies have suggested that there is a ‘core microbiota’ that is particular to specific crops and that is inherited through generations. Targeted manipulation of crop microbiomes with the inoculation of synthetic communities (SynCom) seems to be a promising tool to develop sustainable crop production in the future. In this work, Zhang et al. determined that maize has different core microbiomes in different compartments, with the one from the xylem being most conserved. 25 Operational Taxonomic Units (OTUs) were found in all the xylem sap samples and the culturable ones were able to fix nitrogen (Biological N-fixation, BNF). This finding was verified by the use of GFP-tagged strains for the colonization of the xylem sap and stem and by the use of the 15N isotopic dilution method. In conclusion, maize xylem sap core microbiota is conserved across genotypes and environmental conditions and it contributes to plant N-nutrition through BNF. Understanding core microbiota is a promising tool to enhance crop performance in sustainable agriculture. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Comms. 10.1038/s41467-022-31113-w
Plants and microorganisms build complex associations that are essential for plant homeostasis. Studies have suggested that there is a ‘core microbiota’ that is particular to specific crops and that is inherited through generations. Targeted manipulation of crop microbiomes with the inoculation of synthetic communities (SynCom) seems to be a promising tool to develop sustainable crop production in the future. In this work, Zhang et al. determined that maize has different core microbiomes in different compartments, with the one from the xylem being most conserved. 25 Operational Taxonomic Units (OTUs) were found in all the xylem sap samples and the culturable ones were able to fix nitrogen (Biological N-fixation, BNF). This finding was verified by the use of GFP-tagged strains for the colonization of the xylem sap and stem and by the use of the 15N isotopic dilution method. In conclusion, maize xylem sap core microbiota is conserved across genotypes and environmental conditions and it contributes to plant N-nutrition through BNF. Understanding core microbiota is a promising tool to enhance crop performance in sustainable agriculture. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Comms. 10.1038/s41467-022-31113-w
A boost to plant transformation: Agrobacterium expressing type III secretion system
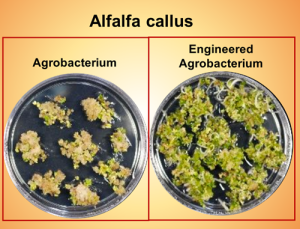 Gene manipulation in most plant species faces two major hurdles, transformation efficiency of tissue and regeneration of callus. Agrobacterium-mediated transformation is the most common method of plant transformation. Transformation efficiency can range from more than half the tissue to less than 1% depending on the types of tissue and plant species. Many times this low efficiency is due to immune responses against Agrobacterium activated by the plant. Since Agrobacterium lacks a type 3 secretion system (T3SS), it cannot effectively suppress immune responses of most plant species. Recently Raman et al. engineered Agrobacterium expressing functional Pseudomonas syringae T3SS and its secreted effectors, and showed that engineered Agrobacterium can deliver effector molecules into the plant cells and suppress plant immune responses. Engineered Agrobacterium shows a significant increase in transformation efficiency in different plant species including the model plants Arabidopsis and Nicotiana benthamiana and the relatively hard-to-transform species wheat, alfalfa, and switchgrass. This work opens a new window to improve genetic engineering in crops and can also help with transgene-free transformation since one can deliver Cas9 via T3SS. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Comms. 10.1038/s41467-022-30180-3
Gene manipulation in most plant species faces two major hurdles, transformation efficiency of tissue and regeneration of callus. Agrobacterium-mediated transformation is the most common method of plant transformation. Transformation efficiency can range from more than half the tissue to less than 1% depending on the types of tissue and plant species. Many times this low efficiency is due to immune responses against Agrobacterium activated by the plant. Since Agrobacterium lacks a type 3 secretion system (T3SS), it cannot effectively suppress immune responses of most plant species. Recently Raman et al. engineered Agrobacterium expressing functional Pseudomonas syringae T3SS and its secreted effectors, and showed that engineered Agrobacterium can deliver effector molecules into the plant cells and suppress plant immune responses. Engineered Agrobacterium shows a significant increase in transformation efficiency in different plant species including the model plants Arabidopsis and Nicotiana benthamiana and the relatively hard-to-transform species wheat, alfalfa, and switchgrass. This work opens a new window to improve genetic engineering in crops and can also help with transgene-free transformation since one can deliver Cas9 via T3SS. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Comms. 10.1038/s41467-022-30180-3