Plant Science Research Weekly: January 24
Review. Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control
 We have only recently begun to appreciate the phenomenon of cross-species or cross-kingdom small RNA transfer, and its applications. Using examples from plants and animals, Cai et al. summarize how some pathogens have evolved the capacity to introduce small RNAs into their host to suppress host defense responses, for example by hijacking host small RNA machinery (e.g., AGO proteins). Conversely, there is also evidence that some plants can secrete extracellular vesicles carrying small RNAs that target pathogen virulence genes. This review also describes the possible applications of small RNAs in plant protection. For example, a plant could be engineered to produce an anti-virulence small RNA, as in host-induced gene silencing (HIGS). Or, to avoid the need for genetic modification of plants, the small RNAs could be provided exogenously, as in spray-induced gene silencing (SIGS), perhaps with the small RNAs being stabilized on clay nanosheets. (Summary by Mary Williams) PLOS Pathogens 10.1371/journal.ppat.1008090
We have only recently begun to appreciate the phenomenon of cross-species or cross-kingdom small RNA transfer, and its applications. Using examples from plants and animals, Cai et al. summarize how some pathogens have evolved the capacity to introduce small RNAs into their host to suppress host defense responses, for example by hijacking host small RNA machinery (e.g., AGO proteins). Conversely, there is also evidence that some plants can secrete extracellular vesicles carrying small RNAs that target pathogen virulence genes. This review also describes the possible applications of small RNAs in plant protection. For example, a plant could be engineered to produce an anti-virulence small RNA, as in host-induced gene silencing (HIGS). Or, to avoid the need for genetic modification of plants, the small RNAs could be provided exogenously, as in spray-induced gene silencing (SIGS), perhaps with the small RNAs being stabilized on clay nanosheets. (Summary by Mary Williams) PLOS Pathogens 10.1371/journal.ppat.1008090
Review: Harnessing atmospheric nitrogen for cereal crop production ($)
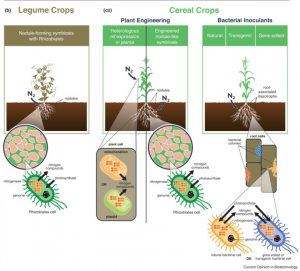 Nitrogen demands for plant growth are accomplished through fertilizers or biological nitrogen fixation. Industrial production of nitrogen fertilizer is expensive and causes pollution due to leaching of unused fertilizer. In this review, Bloch et al. discuss the current status of biological nitrogen fixation and the prospects of biological nitrogen fixation in cereal crops like maize. Leguminous crops like soybean can form a symbiotic relationship with nitrogen-fixing bacteria in the soil to supplement nitrogen demand. Early ancestral cereals were recently shown to associate itself with nitrogen-fixing bacteria. Efforts are being taken to enhance the biological nitrogen fixation in cereal crops by a) engineering plants to be able to form a symbiosis with nitrogen-fixing bacteria, and b) identificating and utilizating nitrogen-fixing bacteria to supplement plant nitrogen requirement. Using molecular approaches like synthetic biology, gene editing, genetics and recombinant DNA technology, multiple research labs are progressing towards this goal. The authors suggest that by the year 2050, much progress will be achieved towards sustainable ways to engineer bacteria that could enhance nitrogen fixation. The adaptability of these rhizosphere bacteria to the natural environment and meeting the regulations apart from managing efficient fertilizer usage will be critical in achieving this. (Summary by Suresh Damodaran) Curr. Opin. Biotechnol. 10.1016/j.copbio.2019.09.024
Nitrogen demands for plant growth are accomplished through fertilizers or biological nitrogen fixation. Industrial production of nitrogen fertilizer is expensive and causes pollution due to leaching of unused fertilizer. In this review, Bloch et al. discuss the current status of biological nitrogen fixation and the prospects of biological nitrogen fixation in cereal crops like maize. Leguminous crops like soybean can form a symbiotic relationship with nitrogen-fixing bacteria in the soil to supplement nitrogen demand. Early ancestral cereals were recently shown to associate itself with nitrogen-fixing bacteria. Efforts are being taken to enhance the biological nitrogen fixation in cereal crops by a) engineering plants to be able to form a symbiosis with nitrogen-fixing bacteria, and b) identificating and utilizating nitrogen-fixing bacteria to supplement plant nitrogen requirement. Using molecular approaches like synthetic biology, gene editing, genetics and recombinant DNA technology, multiple research labs are progressing towards this goal. The authors suggest that by the year 2050, much progress will be achieved towards sustainable ways to engineer bacteria that could enhance nitrogen fixation. The adaptability of these rhizosphere bacteria to the natural environment and meeting the regulations apart from managing efficient fertilizer usage will be critical in achieving this. (Summary by Suresh Damodaran) Curr. Opin. Biotechnol. 10.1016/j.copbio.2019.09.024
Gene duplication accelerates the pace of protein gain and loss from plant organelles
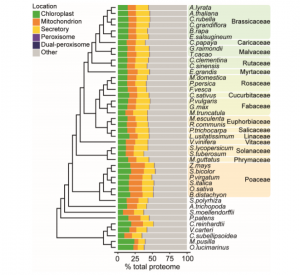 Organelles, such as the chloroplast and nucleus, are structures with specific functions within a plant cell. It has been reported that many related, or homologous, proteins function in different organelles. However, how and why organellar proteins have diverged over evolutionary time remains unclear. Here, Costello et al. find that there has been continual change in the nuclear encoded proteome of organelles, by comparing the protein coding genes of 42 diverse plant species. These changes occur predominantly to genes associated with regulation as well as those involved in stress, development and hormone production. Further analysis identifies a higher rate of change of protein targeting after duplication, supporting the importance of gene and genome duplication for organelle evolution. (Summary by Alex Bowles) Mol. Biol. Evol. 10.1093/molbev/msz275
Organelles, such as the chloroplast and nucleus, are structures with specific functions within a plant cell. It has been reported that many related, or homologous, proteins function in different organelles. However, how and why organellar proteins have diverged over evolutionary time remains unclear. Here, Costello et al. find that there has been continual change in the nuclear encoded proteome of organelles, by comparing the protein coding genes of 42 diverse plant species. These changes occur predominantly to genes associated with regulation as well as those involved in stress, development and hormone production. Further analysis identifies a higher rate of change of protein targeting after duplication, supporting the importance of gene and genome duplication for organelle evolution. (Summary by Alex Bowles) Mol. Biol. Evol. 10.1093/molbev/msz275
Mitochondrial fostering: the mitochondrial genome may play a role in plant orphan gene evolution
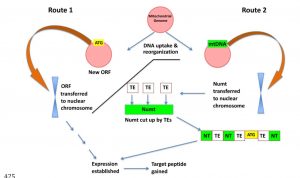 Orphan genes are those that are found in only a single species. In trying to understand the origin of orphan genes, O’Conner and Li have found that many of these orphan genes are likely to have originated as mitochondrial genes, as many are nuclear genes whose encoded proteins are targeted to the mitochondria through encoded targeting peptides. Although mitochondria are derived from bacteria, they retain few genes, with most of their genes having migrated to the nucleus (as much as 98% of mitochondrial genomes can be non-genic). Interestingly, most of the 30 Arabidopsis orphan genes encoded in the mitochondrial genome are also present in the nucleus, further supporting the model that many orphan genes are mitochondrial in origin. Interestingly, mitochondrial genomes have high rates of rearrangement, which could generate the novelty that creates orphan genes. Due to the role of mitochondria in cultivating orphan genes, the authors describe this effect as mitochondrial fostering. (Summary by Mary Williams) bioRxiv
Orphan genes are those that are found in only a single species. In trying to understand the origin of orphan genes, O’Conner and Li have found that many of these orphan genes are likely to have originated as mitochondrial genes, as many are nuclear genes whose encoded proteins are targeted to the mitochondria through encoded targeting peptides. Although mitochondria are derived from bacteria, they retain few genes, with most of their genes having migrated to the nucleus (as much as 98% of mitochondrial genomes can be non-genic). Interestingly, most of the 30 Arabidopsis orphan genes encoded in the mitochondrial genome are also present in the nucleus, further supporting the model that many orphan genes are mitochondrial in origin. Interestingly, mitochondrial genomes have high rates of rearrangement, which could generate the novelty that creates orphan genes. Due to the role of mitochondria in cultivating orphan genes, the authors describe this effect as mitochondrial fostering. (Summary by Mary Williams) bioRxiv
The origin of land plants is rooted in two bursts of genomic novelty
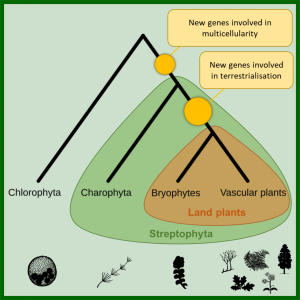 The transition of plants from water to land is one of the most momentous shifts in the history of life on Earth. 500 million years ago, the first land plants dramatically changed the environments on the planet, creating soils, rivers and the oxygen-rich atmosphere. However, the factors that enabled early plants to make the move on to land are not fully understood. Here, by comparing the complete gene sets of over 200 genomes, Bowles et al. demonstrate that two bursts of new genes helped plants make the move from water to land. These genetic innovations enabled the first land plants to protect their embryo, tolerate a range of environmental stresses and anchor themselves to the land. The origin of plants, which now dominate dry environments, was driven by the emergence of genes not seen in close algal relatives (Summary by Alex Bowles) Curr. Biol. 10.1016/j.cub.2019.11.090
The transition of plants from water to land is one of the most momentous shifts in the history of life on Earth. 500 million years ago, the first land plants dramatically changed the environments on the planet, creating soils, rivers and the oxygen-rich atmosphere. However, the factors that enabled early plants to make the move on to land are not fully understood. Here, by comparing the complete gene sets of over 200 genomes, Bowles et al. demonstrate that two bursts of new genes helped plants make the move from water to land. These genetic innovations enabled the first land plants to protect their embryo, tolerate a range of environmental stresses and anchor themselves to the land. The origin of plants, which now dominate dry environments, was driven by the emergence of genes not seen in close algal relatives (Summary by Alex Bowles) Curr. Biol. 10.1016/j.cub.2019.11.090
A single light-responsive sizer can control multiple-fission cycles in Chlamydomonas
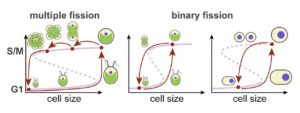 How do cells know when it is time to divide? Helt et al. explore this question using the single-celled alga Chlamydomonas. Unlike most animal and fungal cells, which tend to maintain a relatively consistent size by dividing after their size has doubled, Chlamydomonas cells can undergo several rounds of size doubling during daylight, followed by several rapid divisions (S, M and cytokinesis phases, in what is called a multiple-fission cycle) to bring them back to their standard size. This growth pattern might allow the cells to prioritize photosynthesis and growth when light is available. Several hypothesis have been suggested to explain this pattern, including control by a timer and control by a size criterion. Through experiments and modeling, the authors propose that the multiple-fission cycle can be explained through a molecular “sizer” that measures cell size by its concentration within the cell and that has two regulatory thresholds. With cell volume as its input, the sizer produces a bistable, switch-like response in TF activity and oscillator activity that can account for binary and multiple fission responses.(Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.12.026
How do cells know when it is time to divide? Helt et al. explore this question using the single-celled alga Chlamydomonas. Unlike most animal and fungal cells, which tend to maintain a relatively consistent size by dividing after their size has doubled, Chlamydomonas cells can undergo several rounds of size doubling during daylight, followed by several rapid divisions (S, M and cytokinesis phases, in what is called a multiple-fission cycle) to bring them back to their standard size. This growth pattern might allow the cells to prioritize photosynthesis and growth when light is available. Several hypothesis have been suggested to explain this pattern, including control by a timer and control by a size criterion. Through experiments and modeling, the authors propose that the multiple-fission cycle can be explained through a molecular “sizer” that measures cell size by its concentration within the cell and that has two regulatory thresholds. With cell volume as its input, the sizer produces a bistable, switch-like response in TF activity and oscillator activity that can account for binary and multiple fission responses.(Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2019.12.026
Isolation of an archaeon at the prokaryote–eukaryote interface
 Sometime around 1.8 to 2 billion years ago, complex eukaryotic cells appeared for the first time, providing the ancestor for plants, animals and fungi. Many lines of evidence have indicated that this event probably involved an ancient archaeon taking up an ancient bacterium, the progenitor of all mitochondria. Genomic studies point to the ancient archaeon as belonging to a group of archaea known as the Asgard lineage (within this lineage subgroups are named for Norse gods, and Asgard is the mythical home of Norse gods; hence Asgard lineage). Starting with deep-sea sediments, and after more than ten years of enrichment culturing, Imachi et al. have now successfully cultured an Asgard archaeon. Its growth requires a syntrophic (cross-feeding) hydrogen-scavenging bacterium in co-culture. Study of this syntrophic co-culture led the authors to propose an “entangle–engulf–endogenize” model for the origin of eukaryotic cells. (Summary by Mary Williams) Nature 10.1038/s41586-019-1916-6
Sometime around 1.8 to 2 billion years ago, complex eukaryotic cells appeared for the first time, providing the ancestor for plants, animals and fungi. Many lines of evidence have indicated that this event probably involved an ancient archaeon taking up an ancient bacterium, the progenitor of all mitochondria. Genomic studies point to the ancient archaeon as belonging to a group of archaea known as the Asgard lineage (within this lineage subgroups are named for Norse gods, and Asgard is the mythical home of Norse gods; hence Asgard lineage). Starting with deep-sea sediments, and after more than ten years of enrichment culturing, Imachi et al. have now successfully cultured an Asgard archaeon. Its growth requires a syntrophic (cross-feeding) hydrogen-scavenging bacterium in co-culture. Study of this syntrophic co-culture led the authors to propose an “entangle–engulf–endogenize” model for the origin of eukaryotic cells. (Summary by Mary Williams) Nature 10.1038/s41586-019-1916-6
SUPPRESSOR OF MAX2 1-LIKE 5 promotes secondary phloem formation during radial stem growth
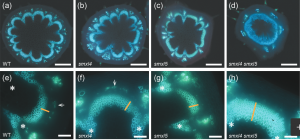 Plant tissues continue to develop postembryonically through proliferation at the shoot and root apical meristem. In the vasculature, radial growth occurs through the proliferation of the undifferentiated cambial cells. Members of the SUPPRESSOR OF MAX2 1-LIKE (SMXL) proteins are known to be involved in primary phloem development in the root vasculature. In this paper, Wallner et al. identified the role of SMXL4 and SMXL5 in secondary phloem formation in Arabidopsis shoot vasculature. SMXL4 is expressed in the differentiated phloem cells and SMXL5 is expressed in the cambium as well as in the mature phloem cells. Individual loss of function alleles of smxl4 and smxl5 showed no changes in the secondary vascular development but the double mutant smxl4 smxl5 had increased cambium-derived tissue. Through loss-of-function mutants and grafting experiments, the authors identified the role of SMXL5 in secondary phloem development. Transcriptomic analysis of smxl4 smxl5 showed downregulation of phloem regulators and the upregulation of stress-responsive genes. Further analysis of phloem marker expression in the mutants indicated the role of SMXL5 in secondary phloem development in the Arabidopsis shoot. Thus SMXL5 has been identified as a regulator of phloem development and a potential key player involved in plant biomass formation. (Summary by Suresh Damodaran) Plant J. 10.1111/tpj.14670
Plant tissues continue to develop postembryonically through proliferation at the shoot and root apical meristem. In the vasculature, radial growth occurs through the proliferation of the undifferentiated cambial cells. Members of the SUPPRESSOR OF MAX2 1-LIKE (SMXL) proteins are known to be involved in primary phloem development in the root vasculature. In this paper, Wallner et al. identified the role of SMXL4 and SMXL5 in secondary phloem formation in Arabidopsis shoot vasculature. SMXL4 is expressed in the differentiated phloem cells and SMXL5 is expressed in the cambium as well as in the mature phloem cells. Individual loss of function alleles of smxl4 and smxl5 showed no changes in the secondary vascular development but the double mutant smxl4 smxl5 had increased cambium-derived tissue. Through loss-of-function mutants and grafting experiments, the authors identified the role of SMXL5 in secondary phloem development. Transcriptomic analysis of smxl4 smxl5 showed downregulation of phloem regulators and the upregulation of stress-responsive genes. Further analysis of phloem marker expression in the mutants indicated the role of SMXL5 in secondary phloem development in the Arabidopsis shoot. Thus SMXL5 has been identified as a regulator of phloem development and a potential key player involved in plant biomass formation. (Summary by Suresh Damodaran) Plant J. 10.1111/tpj.14670
A proposed new classification scheme for fungal and oomycete pathogens based on carbohydrate-active enzymes
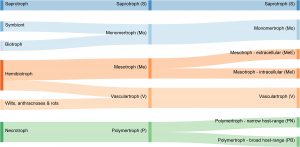 Filamentous pathogens (fungi and oomycetes) use a variety of tactics to obtain nutrients from plants. Classically, they have been categorized as biotrophic (“eating” living tissues), nectrotrophic (eating dead tissues) or hemibiotrophic (biotrophic followed by heterotrophic). Hane et al. point out that these categories are imperfect and difficult to asssign and propose a new approach, based on the organisms’ suite of carbohydrate-active enzymes (CAZymes). The rationale is that these enzymes break down carbohydrates including the cell wall, and should reveal something about how the pathogens access nutrients. Based on this approach, the authors argue for new and additional categories for these pathogens. (Summary by Mary Williams) Frontiers Microbiol. 10.3389/fmicb.2019.03088
Filamentous pathogens (fungi and oomycetes) use a variety of tactics to obtain nutrients from plants. Classically, they have been categorized as biotrophic (“eating” living tissues), nectrotrophic (eating dead tissues) or hemibiotrophic (biotrophic followed by heterotrophic). Hane et al. point out that these categories are imperfect and difficult to asssign and propose a new approach, based on the organisms’ suite of carbohydrate-active enzymes (CAZymes). The rationale is that these enzymes break down carbohydrates including the cell wall, and should reveal something about how the pathogens access nutrients. Based on this approach, the authors argue for new and additional categories for these pathogens. (Summary by Mary Williams) Frontiers Microbiol. 10.3389/fmicb.2019.03088
Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics
 Previously, crop-associated mycobiomes were thought to assemble largely under the control of deterministic selection by the plant host with limited influence from drift. This study by Gao et al., highlights the important role of stochastic processes in fungal community assembly, particularly in the hosts’ early developmental stages and following drought stress. They planted 18 plots of sorghum in a random block design of two host genotypes and three treatments (control, pre-flowering drought, and post-flowering drought) in triplicate and sampled the mycobiome of each of four plant compartments (leaf, root, rhizosphere, and soil) daily using the fungal rDNA ITS2 region, fungal 18S qPCR, and the fungal transcriptome. They hypothesized that drift would most strongly influence community assembly when 1) communities are small and 2) have recently been relieved of selection caused by a stressful event, in this case drought. They found a negative correlation between fungal community size and stochasticity in assembly, supporting H1, but no increase in stochasticity following drought relief, rejecting H2. Understanding the dynamics of fungal mycobiome assembly especially while under drought stress is important in a world where crops will increasingly be threatened by climate change. (Summary by Rebecca Hayes) Nature Comms. 10.1038/s41467-019-13913-9
Previously, crop-associated mycobiomes were thought to assemble largely under the control of deterministic selection by the plant host with limited influence from drift. This study by Gao et al., highlights the important role of stochastic processes in fungal community assembly, particularly in the hosts’ early developmental stages and following drought stress. They planted 18 plots of sorghum in a random block design of two host genotypes and three treatments (control, pre-flowering drought, and post-flowering drought) in triplicate and sampled the mycobiome of each of four plant compartments (leaf, root, rhizosphere, and soil) daily using the fungal rDNA ITS2 region, fungal 18S qPCR, and the fungal transcriptome. They hypothesized that drift would most strongly influence community assembly when 1) communities are small and 2) have recently been relieved of selection caused by a stressful event, in this case drought. They found a negative correlation between fungal community size and stochasticity in assembly, supporting H1, but no increase in stochasticity following drought relief, rejecting H2. Understanding the dynamics of fungal mycobiome assembly especially while under drought stress is important in a world where crops will increasingly be threatened by climate change. (Summary by Rebecca Hayes) Nature Comms. 10.1038/s41467-019-13913-9
From population to production: 50 years of scientific literature on how to feed the world
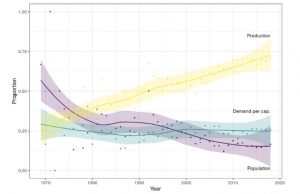 Tamburino et al. analyzed text from more than 12,000 research articles published in the past 50 years that included the terms “global” or “world” and “food supply”, “food demand’, or “zero hunger”. From this dataset, they quantified terms related to population, total food production, or per-capita demand (e.g., through different types of diets). Across this time span, the authors found a trend for a decreasing focus on population size, and an increasing focus on how to increase food production. Speculating on the cause of these trends, the authors suggest that framing food security in terms of population is politically sensitive, disproportionally blames developing countries, and raises the possibility of coercion as in the one-child policy of China. Furthermore, economic development is already promoting a decrease in rate of population growth. Yet, the authors point out that trends in food production increases suggest they are not great enough to meet demands, particularly with rising levels of meat consumption and the threats of climate change. The authors propose a renewed focus on per capita consumption and research that addresses all three factors at the same time. (Summary by Mary Williams) Global Food Security 10.1016/j.gfs.2019.100346
Tamburino et al. analyzed text from more than 12,000 research articles published in the past 50 years that included the terms “global” or “world” and “food supply”, “food demand’, or “zero hunger”. From this dataset, they quantified terms related to population, total food production, or per-capita demand (e.g., through different types of diets). Across this time span, the authors found a trend for a decreasing focus on population size, and an increasing focus on how to increase food production. Speculating on the cause of these trends, the authors suggest that framing food security in terms of population is politically sensitive, disproportionally blames developing countries, and raises the possibility of coercion as in the one-child policy of China. Furthermore, economic development is already promoting a decrease in rate of population growth. Yet, the authors point out that trends in food production increases suggest they are not great enough to meet demands, particularly with rising levels of meat consumption and the threats of climate change. The authors propose a renewed focus on per capita consumption and research that addresses all three factors at the same time. (Summary by Mary Williams) Global Food Security 10.1016/j.gfs.2019.100346



